Evaluation of the Structure and Conformation of Albumin Protein after Interaction with Graphene and Graphene Oxide
Hossein Arzani1, Fatemeh Ramezani2*
1Department of Medical Physics and Biomedical Engineering, Shahid Beheshti University of Medical Sciences, Tehran, Iran
2Physiology Research Center, Iran University of Medical Sciences, Tehran, Iran
*Corresponding author. E-mail: Ramezani.f@iums.ac.ir
Received: Sep. 07, 2022; Revised: Dec. 02, 2022; Accepted: Dec. 22, 2022; Published: Dec. 31, 2022
Citation: H. Arzani, F. Ramezani. Evaluation of the structure and conformation of albumin protein after interaction with graphene and graphene oxide. Nano Biomedicine and Engineering, 2022, 14(4): 375–384.
DOI: 10.5101/nbe.v14i4.p375-384
Abstract
Given the increasing demand for graphene-based nanomaterials, it seems necessary to study their effects on the biomolecules they encounter. In this study, we will present the results of our computational study on the influence of graphene and graphene oxide (GO) on albumin. The interaction between albumin, graphene and GO was investigated using molecular dynamics and GROMACS software for 100 ns with a TIP3P water model. Binding energy, its root-mean-square deviation and structural changes were calculated. The molecular dynamics results show that the interaction of albumin with both graphene and GO structures causes changes in protein structure and that GO has a larger effect on the secondary structure. The results show that the binding energy between albumin and graphene is higher than that of GO. The analysis shows that it is more related to non-polar or hydrophobic interactions in the albumin-graphene interaction. Graphene and GO affect the protein microenvironment and reduce the amount of secondary structure, but in general the degree of change depends on the ratio of graphene or GO to albumin, GO functional groups, temperature and pH.
Keywords: Graphene; GO; Albumin; Interaction; Secondary structure
Introduction
With the advent of nanotechnology, nanomaterials have shown great potential in the diagnosis and treatment of diseases. Carbon nanomaterial such as graphene [1–8] is a two-dimensional atomic-thickness carbon nanomaterial whose physical and chemical properties make it suitable for a variety of applications, such as defense equipment, cosmetics, textiles, drug delivery and energy storage [9–11].
Concerns on its biocompatibility have increased due to the increasing demand for graphene-based nanomaterials with different sizes and functional groups in the medical field. Accordingly, understanding the relationship and interaction of proteins with graphene is particularly important. Detailed knowledge of protein-graphene complex formation can also be used to develop biological applications of graphene such as more efficient biosensors, drug delivery and protein separation processes. To increase our ability to engineer such complexes, it is essential to understand and preferably be able to predict the adsorption capacity and structure of proteins after adsorption on graphene.
Concerns on its biocompatibility have increased due to the increasing demand for graphene-based nanomaterials with different sizes and functional groups in the medical field. Accordingly, understanding the relationship and interaction of proteins with graphene is particularly important. Detailed knowledge of protein-graphene complex formation can also be used to develop biological applications of graphene such as more efficient biosensors, drug delivery and protein separation processes. To increase our ability to engineer such complexes, it is essential to understand and preferably be able to predict the adsorption capacity and structure of proteins after adsorption on graphene.
Albumin is the most abundant plasma protein at a physiological concentration of 40–50 g/L (usually approximately 50% of human plasma protein), which causes 70% of the plasma oncotic pressure [12–15]. Albumin is a negatively charged plasma protein with an isoelectric point of 4.3–4.9. The outer part of the molecule is composed of hydrophilic amino acids and its center is composed of hydrophobic amino acids that can attach to molecules with low solubility in water and carry drugs and ligands [15–17]. Bovine serum albumin (BSA) and human serum albumin (HSA) have a similar structure and amongst the most widely used proteins in the pharmaceutical field [8]. Given the importance of this protein, its interaction with the nanostructure passing in the blood can be important [18] .
Analysis of these types of interactions between proteins and ligands or nanomaterials is performed in the laboratory by experimental methods such as spectroscopy. Albumin emits fluorescence due to the amino acids tryptophan, tyrosine and phenylalanine. Protein fluorescence is usually affected by the quantity and position of tryptophan because the fluorescence intensity of tryptophan is eight times that of tyrosine and 140 times that of phenylalanine. Thus, tryptophan is the major molecule of the chromosphere. Protein deformation can be detected when interacting with other materials using fluorescence and circular dichroism (CD) spectroscopy. The intrinsic fluorescence spectrum of proteins can be used to study the local environmental change (tertiary protein structure) of these amino acid residues. A possible change in the maximum wavelength propagation indicates a change in polarity around the chromophore molecule[19] . Computer simulations of atomic dimensions, such as molecular dynamics (MD), are also used to study the interaction of proteins with ligands and nanomaterials.
In this study, the effect of graphene and graphene oxide (GO) on the protein structure of albumin was investigated using computational simulations, and the simulation results were compared with previous studies.
Methods
Molecular dynamics
The required structural file for the albumen was obtained from the PDB database. The structural file of graphene and GO was created using a plugin in VMD software. First, the protein files were simulated for a minimum of energy for 10 ns to reach their ideal structure. Then the output coordinate file was used as the input of the next step. The interaction of protein with graphene surface was investigated using AutoDock software to determine the correct direction of bonding between graphene and protein, based on the binding energy. In this software, graphene is allowed to bind to all directions of the protein by considering all the atoms, and finally, based on the binding energy, the most stable bond is considered as the most likely bond. Then, the PDB file of the protein-graphene complex, which had the lowest binding energy, was used as the MD input file. MD simulations were performed using GROMACS software according to several previous studies [20–24]. All bonded and non-bonded parameters for graphene atoms were entered into the OPLSAA force field. All-atom simulation was performed between the graphene sheet and protein for 100 ns. TIP3P water model simulation was performed under neutral NaCl concentrations. The temperature was adjusted to 300℃ and maintained by the NoseHoover thermostat. The pressure was maintained at 1 atmosphere by the Berendsen barostat. Time steps of 2 fs were considered for all simulations. The cut-off of the Vander Waals waves started at a distance of 1.1 nm and reached zero at 1.2 nm by the Ewald mesh method. PME 1.4 nm was used to calculate the electrostatic interactions with a cut-off distance. The original simulation was performed at 100 ns. The root-mean-square deviation (RMSD) and the binding energy were calculated to determine the interaction energy between the proteins, the graphene and GO plates.
To evaluate the stability of the simulation in MD, the properties of each set (such as temperature, pressure, energy and structure) along the entire MD path are examined. One of the most widely used modules in GROMACS to evaluate the stability of systems is g_rms. In this way, the RMSD of the systems can be obtained to compare the stability and suitability of the ligand and the simulation conditions. RMSD values of the backbone of atoms are used to monitor the dynamic stability of MD pathways:
![]() (1)
(1)
To evaluate the intensity of protein interactions with surfaces or ligands and to determine the appropriate reaction conditions, it is very important to calculate the free binding energy, which is calculated based on the following formula:
(Receptor energy + Ligand energy) - Receptor energy and ligand = Free binding energy
ΔGbinding is a thermodynamic value whose negative values indicate better bonding and better ligand conditions. The free energy of the connection is measured using various methods. These include methods for free perturbation energy (FEP), thermodynamic integration (TI), meta-dynamics, umbrella sampling, the linear interaction method, and the Poisson-Boltzmann surface area (MM-PBSA). Easy access and fast and easy calculation of free binding energy with the help of MM-PBSA have led to the increasing use of this method, especially in the biological sciences. In addition, the algorithms of this method are compatible with GROMACS modules.
The g_mmpbsa method was used in this study. In this method, based on the proposed formulas, four energies, including electrostatic and van der Waals, polar and nonpolar forces are calculated. The connection energy is then calculated using MmPbSaStat.py. For this purpose, the last 20 ns of the simulation trajectories were used during 300 snapshots.
![]()
![]() (2)
(2)
![]() (3)
(3)
![]() (4)
(4)
![]() (5)
(5)
The simulation results were analyzed using VMD software (a molecular visualization program used to display, animate, and analyze simulated systems). Other software, such as Viewer lite, Discovery Studio, Ligand scout, Hyperchem, and LAMMPS, were used to prepare the initial file and analyze the results.
Search strategy
For a complete search, we searched for articles on the effect of graphene and GO on albumin structure and function, which were published by MEDLINE, SCOPUS, EMBASE (as of Web of Science, March 31, 2022), without linguistic or temporal limitations. The keywords used to search in PUBMED for graphene and albumin are listed below:
Selection criteria, eligibility and elimination of study and information extraction
After removing duplicate articles, the two researchers screened separately by reading the titles, abstracts, and full text of the articles (if the titles and abstracts did not contain enough information). Disagreements regarding whether the study should be conducted were resolved through discussion. All experimental and computer studies investigating the effect of graphene or GO on albumin protein have been included in the study. Exclusion criteria were as follows: review articles, articles that targeted proteins other than albumin, and articles that did not use graphene or GO as intervention. Both independent researchers extracted the following data from this study: type of study (experimental or computer), type of nanoparticles and type of test.
Results
Protein and graphene docking
The Albumin protein alone was first subjected to 10 nm MD simulation and the final file was used as an input file for docking (Fig. 1). Figure 1(c) shows the most stable graphene-albumin structure (binding energy = -5.68 kJ/mol). These structures have been used as dynamic molecular input files.

Fig.1 Structure of (a) graphene, (b) albumin, (c) albumin-graphene complex due to docking of these two molecules along with the binding energy of docking
Figure 2 shows the interaction of albumin with graphene (Fig. 2(a)) and GO (Fig. 2(b)) , showing the changes made on proteins after 100 ns of simulation. The structure of graphene is more affected by the interaction with protein than that of GO.
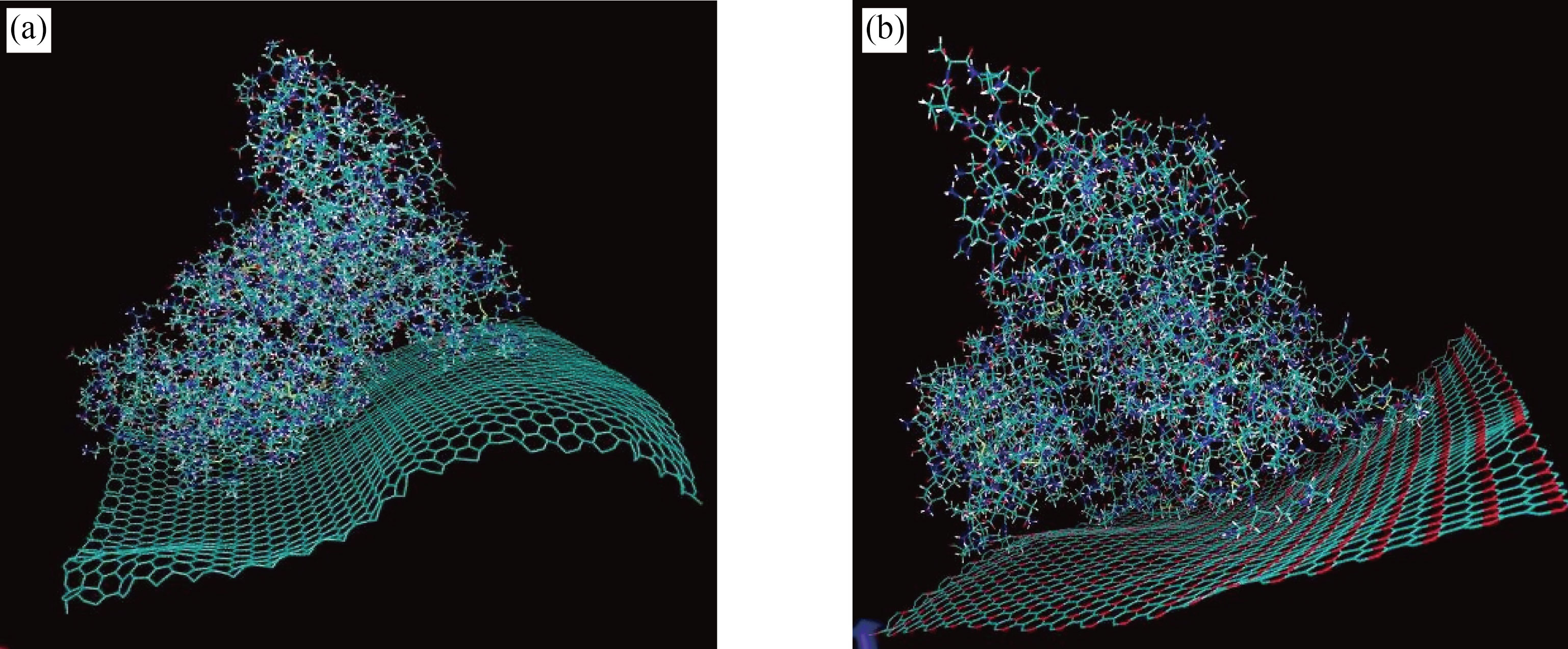
Fig.2 Interaction of albumin with (a) graphene and (b) graphene oxide showing the changes made on plates and proteins after 100 ns of simulation
Changes in the secondary structure of albumin after adsorption on the graphene
Figure 3(a) shows the secondary structure changes in each protein after interaction with graphene and GO. The secondary structure of albumin decreased from 425 to 400 amino acids during interaction with graphene. Alpha-helix also decreased from 350 to 325 amino acids. The number of amino acids in the bend structure increased from 40 to 60 amino acids. Figure 3(b) shows the amount of secondary structure in albumin protein during 100 ns interactions with GO plates. The amount of secondary structure in albumin protein decreased from 425 to 400 units. Alpha-helix also decreased from 350 amino acids to 315 amino acids. The number of amino acids in the coil structure increased from 75 to 100 amino acids.
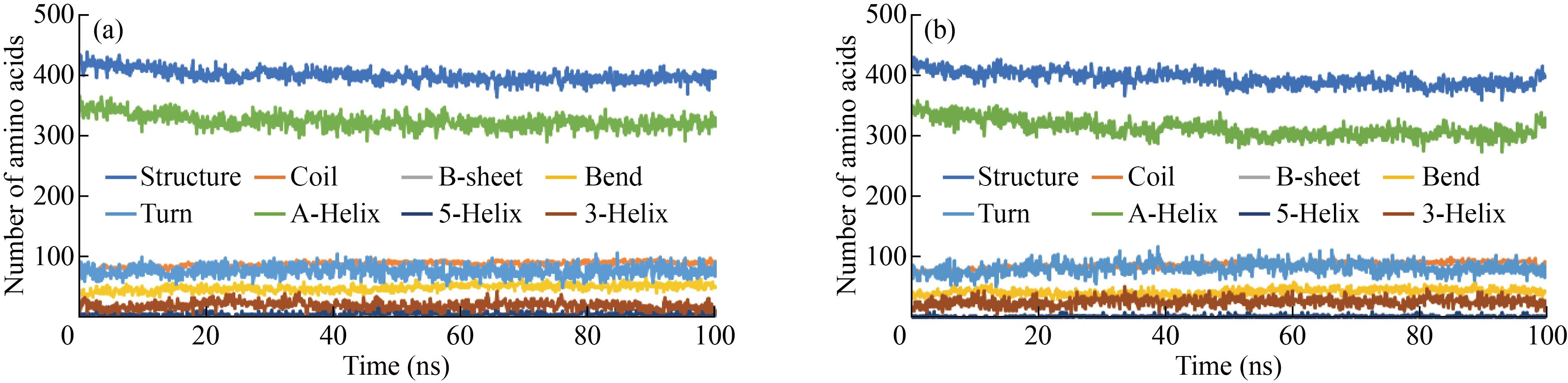
Fig.3 Rate of change in the secondary structure of Albumin after interaction with (a) graphene and (b) graphene oxide
Gyration radius of protein on the graphene and graphene oxide surfaces
The gyrus radius is a parameter that indicates the density and stability of the structure (Fig. 4). This value has changed significantly in the studied models. The average radius of the graphene-albumin model during the simulation is 2.56 nm, while this amount increased in the graphene oxide-albumin model, and this factor probably caused more openings and changes in the structure of the albumin.
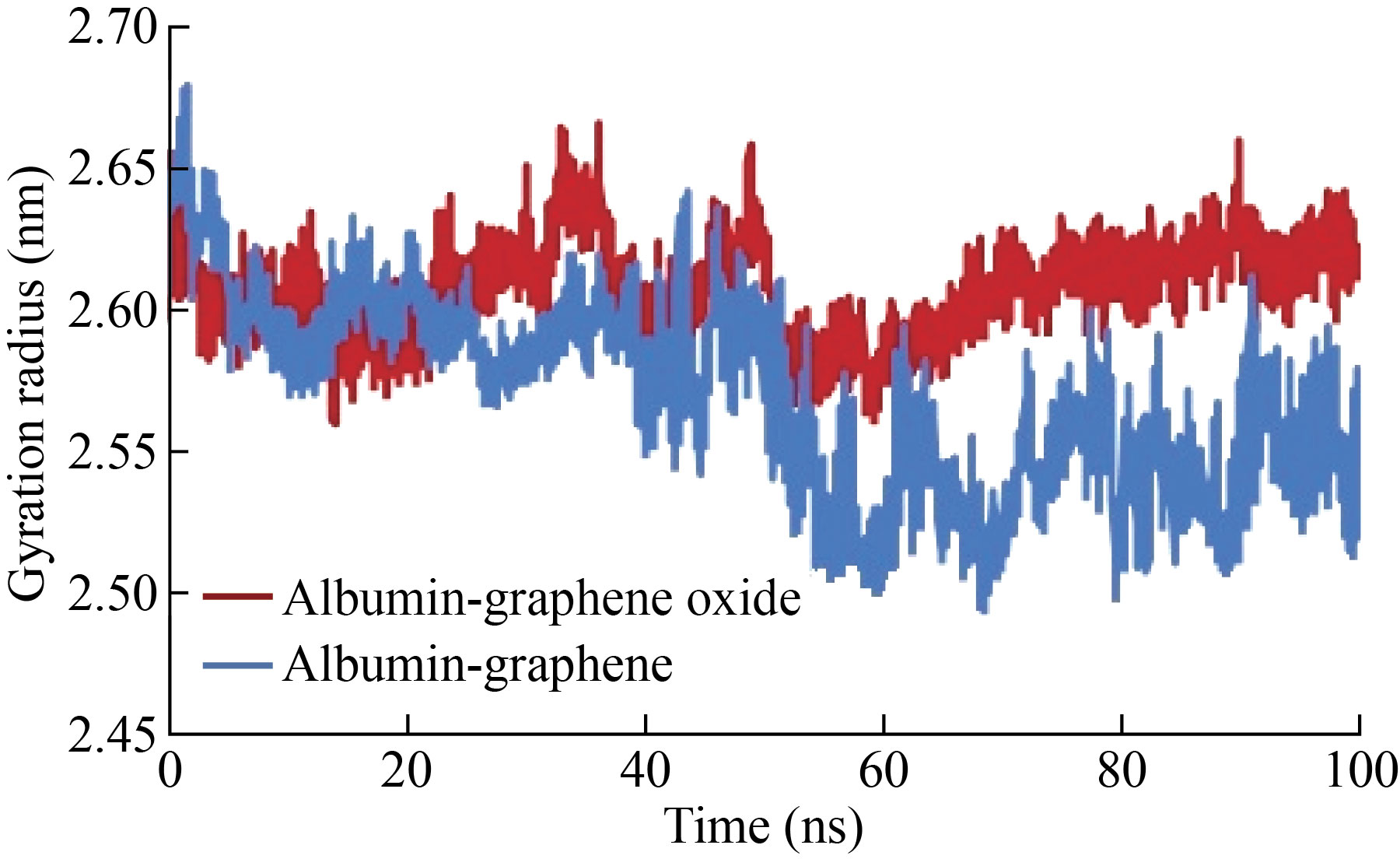
Fig.4 Gyration radius of albumin during interaction with graphene and graphene oxide
Comparison of protein RMSD on graphene and graphene oxide surfaces
Figure 5 shows that the RMSD of the protein exposed to the surface of graphene and GO is different, so the amount of these deviation in the albumin-GO exposure is greater than the deviation in the exposure of this protein to graphene. Based on this, the average deviation of albumin-GO was equal to 0.45 nm and the average of this parameter in albumin-graphene was 0.33 nm.
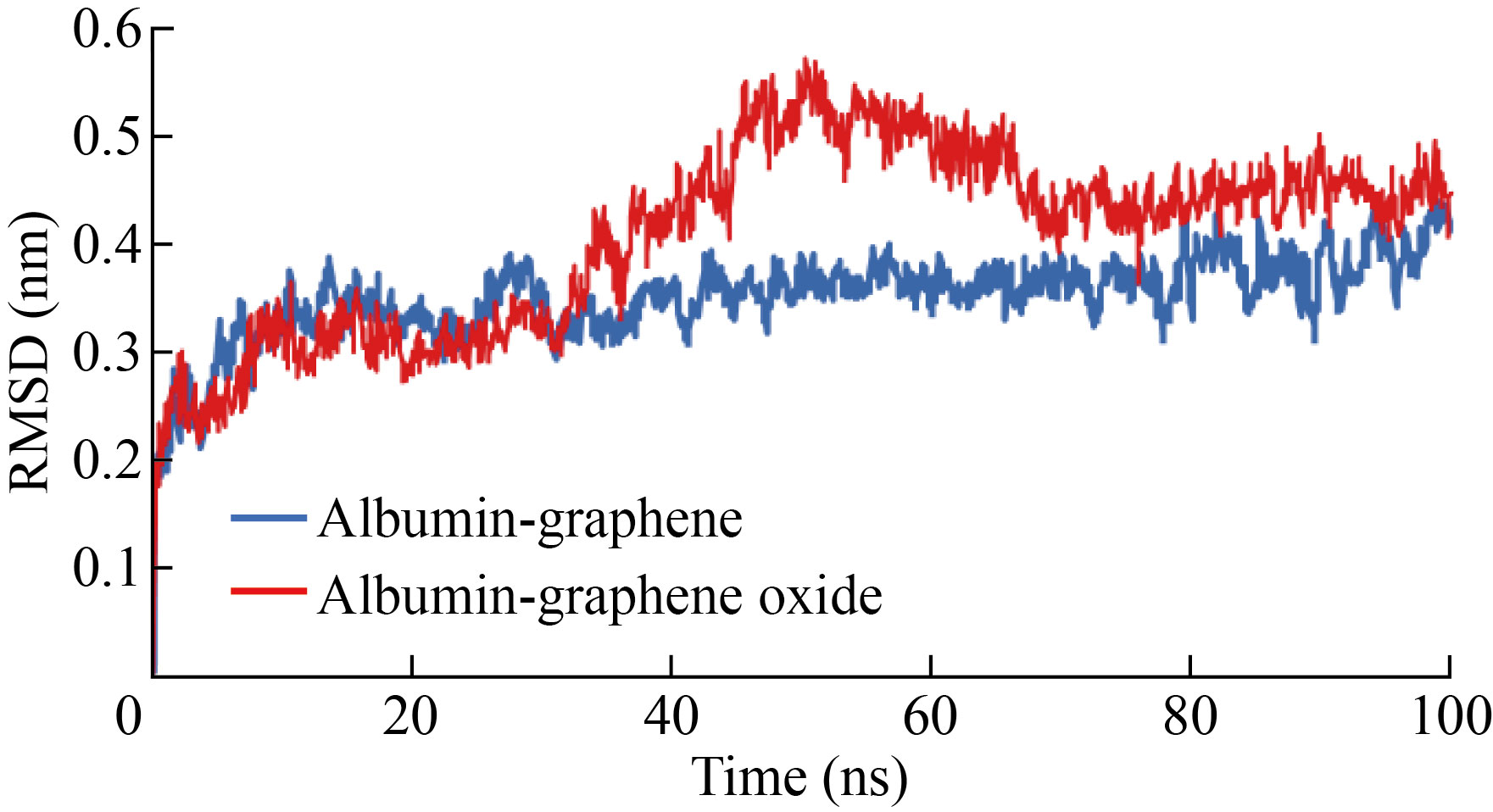
Fig.5 RMSD of albumin protein (high) on the surface of graphene and graphene oxide
Changes in the number of hydrogen bonds in the protein structure on the surface of graphene and graphene oxide
One of the important parameters in determining the structural stability of proteins and the interaction between surfaces and other proteins is changed in the number of hydrogen bonds. As shown in Fig. 6, the number of hydrogen bonds of albumin exposed to graphene oxide was greater than that of albumin exposed to graphene. In other words, the internal and structural changes of the protein in the case of exposure to graphene oxide were probably less. The average number of graphene-albumin bonds during the simulation was 417, and the average number of hydrogen bonds of albumin-GO was 425.
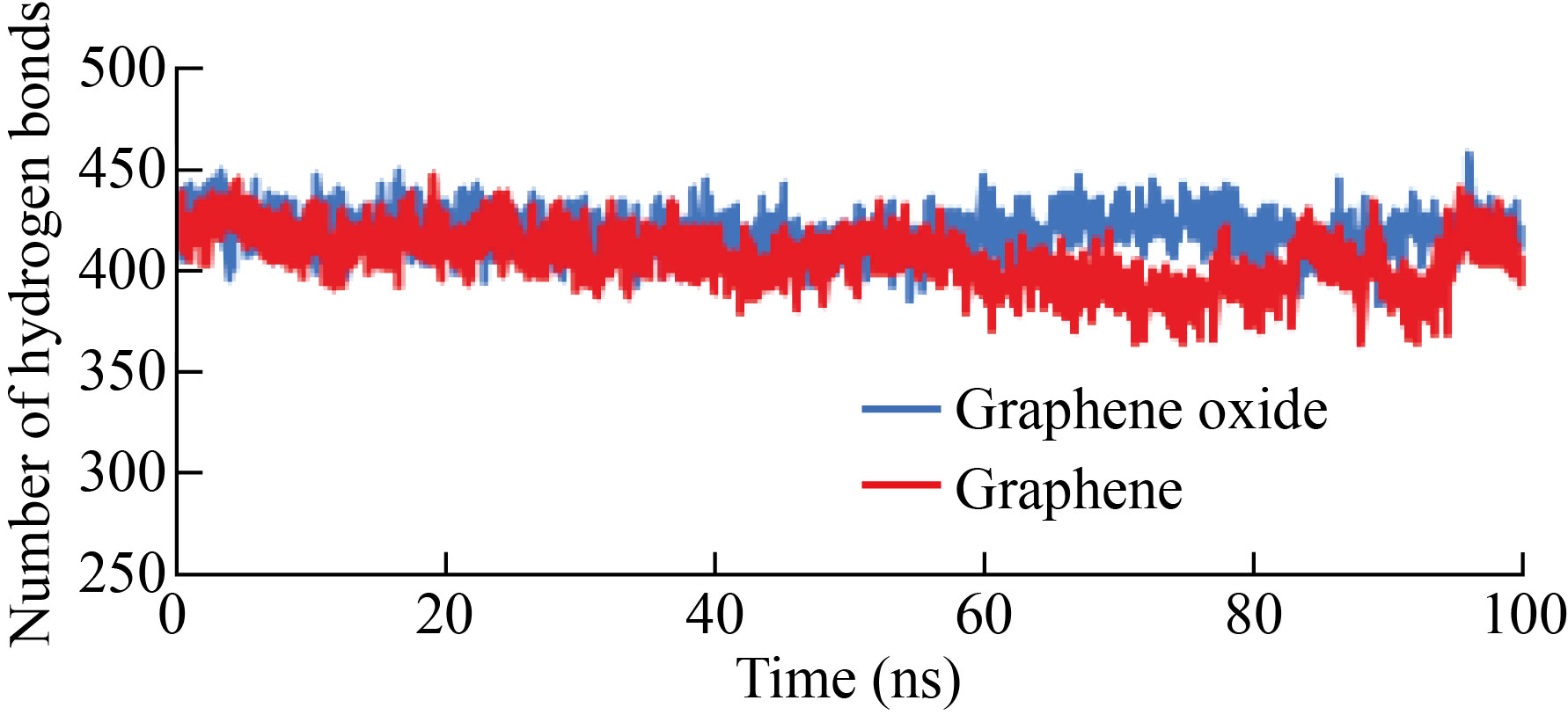
Fig.6 Number of hydrogen bonds of albumin protein on the surface of graphene and graphene oxide
Investigation of protein binding energy to graphene and graphene oxide surface
Table 1 shows the van der Waals, electrostatic, nonpolar and polar binding energy, solvent pressure proportionality (SAV), and ΔG of binding of graphene and GO to albumin. The results show that the binding energy between graphene and albumin (-750.025 kJ/mol) is higher than that between GO (-524.500 kJ/mol). Although the amount of van der Waals and electrostatic interactions in the albumin-GO interaction is higher than these parameters in the albumin-graphene interaction, nonpolar interactions are much higher in the albumin-graphene interaction.
Table 1 Amount of energy released due to the interaction of graphene and graphene oxide with albumin
ΔGbinding (kJ/mol) | Non-polar (SAV model) (kJ/mol) | Non-polar (SASA model) (kJ/mol) | Polar solvation energy (kJ/mol) | Electrostatic (kJ/mol) | van der waals (kJ/mol) | Energy |
-750.025 | -596.315 | -68.694 | 932.976 | 0 | 1017.993- | Albumin-graphene |
-524.500 | -263.627 | -56.758 | 1326.056 | 12.23- | 1518.178- | Albumin-graphene oxide |
Results of Systematic Review
A total of 2968 articles were obtained from extensive database searches. After removing duplicate articles, a total of 2374 articles remained. A total of 211 articles were selected based on title evaluation. After evaluating the full texts, 15 articles containing sufficient information were selected for systematic review. Figure 7 shows the flow chart of the article search and selection process. Fourteen studies investigated the effect of graphene and graphene derivatives on the albumin structure. The information obtained from the articles is summarized in Table 2. Twelve articles reviewed the effect of GO, two reviewed the effect of graphene quantum dot (GQD), and one reviewed the effect of graphene. Six studies were on HSA, Six studies on BSA, and two studies on both. In one study, the type of albumin was not definite. Except for one MD study, the rest of the studies were performed in the laboratory.
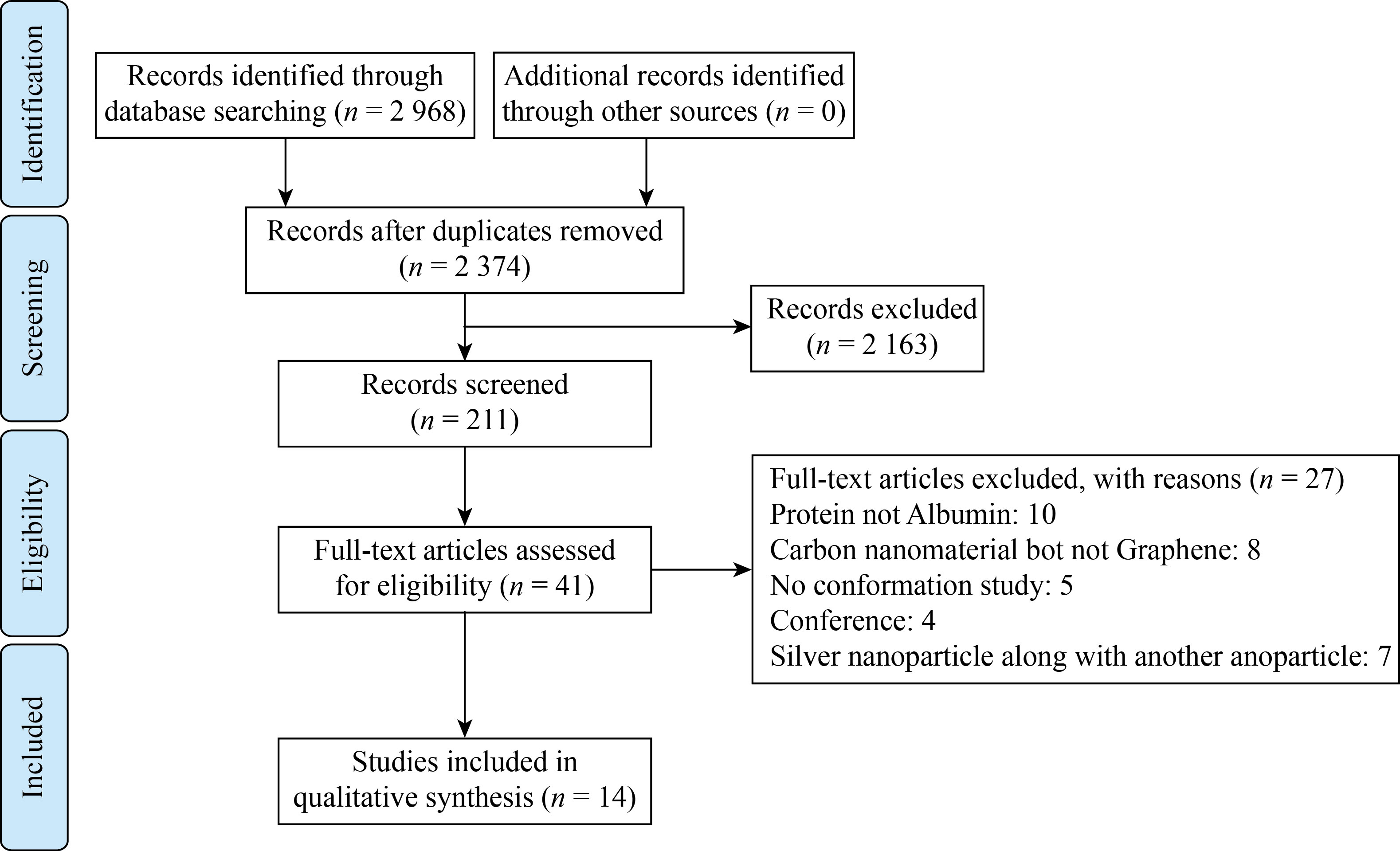
Fig.7 PRISMA flow chart for a systematic overview showing the details of the database search, the number of abstracts and the full text of the study
Table 2 Data extracted from included articles
Outcomes | Method of investigation | Nanoparticle/Protein | Reference |
(1) Ka values at 298 K are equal to 6.46×105 L·mol−1 (2) The fluorescence quenching is static quenching (3) Reactions between GQDs and HSA are driven by hydrogen and van der Waals bond (4) GQDs had a negligible effect on the secondary structures of HSA | Experiment | GQD/HSA | [25] |
(1) The driving forces of the binding include covalent, hydrogen, electrostatic bonds, hydrophobic interactions and p–p stacking effects (2) GO may make conformational changes in HSA and failure in HSA's binding ability to toxins and prevent the function of HSA principally through blocking the protein active site (3) GO-COOH interacts with HSA mainly through hydrogen bonds and displays lowest conformational and functional changes. (4) GO-CS shows similar but slightly worse biocompatibility to HSA (5) GO-PEI almost abolishes the structure and function of HSA through the interruption of protein structure | Experiment | (1) GO (2) GO-COOH (3)GO-CS (4)GO-PEI/HSA | [26] |
The binding of GO-COOH to HSA outcomes in minimal conformational change and HSA’s binding ability to bilirubin remains unaffected, while the binding of p-GO and GO-PEI shows strong toxicity on HSA | Experiment | (1) GO-COOH (2) GO (3) GO-PEI/HSA | [27] |
(1) GO could vary the secondary structures and conformation of BSA and decrease of a-helix content (2) GO initiated the fluorescence quenching of BSA | Experiment | GO/BSA |
[19] |
(1) GQDs could bind and alter the total and local conformational, and function of HSA (2) GQDs could quench the intrinsic fluorescence of HSA via static mode (3) Binding site of GQDs was site I of HSA (4) GQDs interacted with HSA mainly through van der Waals interactions, hydrogen bonding interactions, and protonation | Experiment | GQD/HSA | [28] |
(1) Albumin interacts with GO and GQD via van der waals or hydrogen bonding (2) The amount of secondary structure rises from 58% in native HSA to 53% in the presence of GO | Experiment/ Docking | GO/HSA |
[29] |
(1) GO as a carbocatalyst has nonspecific endopeptidase activity cleavage BSA, but depends on temperature, time, and the amount of GO (2) Cleavage of the proteins did not result in thorough hydrolysis into their principal amino acids | Experiment | GO/BSA | [30]
|
(1) Static quenching of HSA and BSA (2) Different quenching efficiency between GO and Trp or Tyr at different PHs showed the importance of electrostatic interaction in quenching. Hydrophobic interaction also takes part in quenching | Experiment | GO/HSA and BSA | [18] |
(1) Electrostatic interactions has a key role in controlling HSA-GO interactions. Adding Calcium ions also aided HSA adsorption (2) Maximum HSA adsorption was observed at the isoelectric point of HSA (3) Under acidic conditions, the adsorption of HSA on GO led to the formation of protein layers with a high degree of fluidity due to the stretched conformation of HSA | Experiment | GO/HSA | [31] |
(1) Static quenching, combined with dynamic quenching (2) Negative values of enthalpy(∆H), entropy (∆S) and free energy (∆G) change (3) Interaction between BSA and GO was van der Waals interaction or hydrogen bond (4) Quenching process was exothermic and spontaneous (5) Energy transferring from BSA to GO had high chance (6) Conformation of BSA was changed | Experiment | GO/BSA | [32] |
(1) Static quenching in combination with dynamic quenching (2) The best conditions for the most effective affinity are neutral pH and room temperature (3) The strong impact of the size of GO on the interaction between proteins and GO (4) The interaction between GO and albumin is electrostatic and hydrophobic. (5) Decreasing helicity in the secondary structure of albumins | Experiment | GO/HSA and BSA | [33] |
BSA was affected the GO stability concentration dependently | Experiment | GO/BSA | [34] |
No major change in protein structure | Molecular dynamic | Graphene/NR | [35] |
(1) The GO surface yielded higher BSA adsorption than reduced GO (2) The main forces during the adsorption of albumin onto graphene oxide and reduced GO are shown to be hydrophilic and hydrophobic interactions, respectively (3) BSA on GO can hold its binding sites, but a denatured layer of BSA forms on the reduced GO followed by further binding of active BSA molecules, depending on the concentration of the protein | Experiment | GO and reduced GO /BSA | [36] |
Discussion
The simulation results showed that the binding energy between albumin and graphene is higher than that of GO, which is more related to nonpolar interactions in contact with graphene. This finding is confirmed by experimental studies that have reported the dominant forces during the adsorption of albumin on GO and reduced GO are hydrophilic and hydrophobic interactions, respectively [36]. The hydrophilic surface of Graphene oxide containing different oxygen functional groups showed a greater tendency than graphene to absorb protein. Therefore, the amount of albumin adsorbed on the surface of GO was increased more than that of GO. On the other hand, due to the increase in hydrophobicity on the surface of reduced GO, the amount of protein absorbed on it decreases [36]. Thermodynamic studies also showed that the interaction of albumin with GO and GQD occurs through the polar interaction of van der Waals or hydrogen bonding [18, 25, 28, 32, 35].
By investigating the effect of pH, the electrostatic interaction between albumin and GO was also confirmed [31, 33]. In the study of the interaction of albumin and GO at different pH values, it is found that the amount of albumin quenching at pH=7.01 is much higher than that at pH 3.00 and 10.98, which indicates that the tendency of GO bonding to albumin under acidic pH (pH=3.00) and alkaline (pH=10.98) conditions is lower than that under physiological conditions (pH=7.01) [33]. At pH values less than the isoelectric point, albumin has more positive charges. Thus, the positive charge of albumin at pH =7.01 is greater than that at pH=10.98 and at pH=3.00 [26, 28]. Based on this knowledge, it can be expected that the maximum interaction between graphene oxide and albumin occurs at pH 3.00, and with increasing pH, the interaction should decrease, but the experimental results showed that the interaction at pH=3.00 is weaker than that at pH =7.01. This difference is due to structural changes in proteins that increase the hydrophobicity of the protein and affect the electrostatic interactions. In fact, the combination of these two forces determines the table of interactions between the protein and the GO surface. At pH=10.98, a slight quenching of albumin is observed in the presence of GO, which can be explained by negative charges on the surface of albumin, the amount of which increases towards an alkaline pH. Based on these observations, it can be said that the strong quenching of albumin by GO is mainly due to the electrostatic attraction between albumin and GO [33]. Confirmation of the effect of electrostatic forces on the interaction of albumin with graphene oxide surface has shown that increasing the ionic strength (e.g., blood compared to saliva) under neutral pH conditions leads to a stronger bond between albumin and GO as well as the formation of albumin layers. It focuses on GO, which emphasizes the key role of electrostatic interactions in controlling HSA-GO interactions. Calcium ions also likely facilitate albumin uptake through charge neutralization and bridging effect. Ionic and pH conditions as well as the presence of calcium ions had key effects on the binding strength of albumin-GO and the composition of albumin-GO, which indicates the influence of electrostatic bonds on the interaction [31].
The results of MD simulation showed that the secondary structure and especially the amount of alpha helix on both graphene and GO structures decreased. The reduction in the secondary structure of albumin during interaction with GO is greater than when it interacts with graphene. The radius of gravity of albumin on GO is also on average longer than when it interacts with graphene, which indicates more openness and changes in the structure of albumin during interaction with GO.
Static fluorescent quenching of albumin after interaction with GO [18, 19, 28] and GQDs [25] and static quenching combined with dynamic quenching [32, 33] were demonstrated in experimental studies. The results of experimental studies in 9 studies using the fluorescence quenching test and circular duality (CD), confirming GO can alter the secondary structures of albumin ]19–33, 36]. The presence of GO reduces the helical albumin content [19, 33]. According to CD spectroscopy, HSA shows two negative absorption bands of approximately 208 nm and 222 nm, representing the typical helical structure of HSA. The addition of GQDs reduced the bandwidth at all HSA wavelengths, indicating that the interaction of GQDs with HSA leads to structural changes in HSA. The content of the helix decreases but the other contents of the secondary structure increase with increasing concentrations of GQDs, indicating a decrease in the biological activity of HSA and the degradation of HSA hydrogen bond networks in interaction with GQD [28]. FT-IR spectroscopy confirmed that GQDs interact with the C–O, C–N and N–H groups in HSA polypeptides, leading to rearrangement of the HSA structure. The binding site of GQDs is mainly site I of HSA (warfarin binding site) and some common ions can increase the binding capacity of GQDs to some extent [28]. Ding et al. also reported in 2014 that GO easily immobilizes protein and inhibits protein function. GO inhibits HSA function primarily by blocking the active site [26].
Contradictory result was obtained in experimental study performed by Xiao-Xu Ba in 2019 [25]. They showed that GQDs had little effect on HSA secondary structures. GQDs have no additional effect on the polar microenvironment of the amino acids Tyr and Trp in HSA. No obvious changes in the specific peptide band of the protein were observed in the presence of GQDs, and the shape and position of the peaks in the CD spectra did not show any significant change. The percentages of calculating the results of α-helix, β-strand, round and random coils also do not change significantly, meaning that GQDs do not disturb the secondary structure of HSA [25], and the basic structure of proteins after binding to GO is still mainly an alpha helix[33] . It should be noted that the secondary structure of albumin is entirely concentration dependent and the conflicting results may be related to the ratio of GO to albumin. The CD results showed that the content of the helix of HSA before exposure to GO and in the presence of different concentrations of GO was as follows: 58% native HSA, 57.3% (HSA+5 mg/mL graphene oxide), 57% (HSA+10 mg/mL GO), 56% (HSA+25 mg/mL GO) , 55.5% (HSA+30 mg/mL GO) , 54.8% (HSA+35 mg/mL GO), 53% (HSA + 50 mg/ml GO). The amount of secondary structure decreases from 58% in Native HSA to 53% in the presence of GO [29].
In the MD study of the albumin interaction with graphene, it has been reported that despite changes in torsion angle in C–N and albumin skeletons and the appearance of beta sheets, no major changes in albumin conformation have occurred[35] .
When the function of adsorbed BSA in interaction with anti-BSA antibody was examined, it was found that BSA adsorbed on GO could maintain its specific binding site to the antibody. Therefore, despite the adsorption of albumin on the GO surface and its conformational changes, in this study it was reported that no functional changes were observed in albumin, which may be due to the multilayer adsorption of protein on the GO surface and structural changes only in the first layer. Additionally, albumin is denatured after adsorption on reduced GO [36].
In studies of GO derivatives, it was observed, GO-COOH has high biocompatibility with HSA with minimal conformational and functional changes[26] . GO-COOH binding to HSA does not change the binding capacity to bilirubin [27]. GO-CS has a similar but slightly worse biocompatibility to HSA than GO-COOH, while GO-PEI almost destroys the structure and function of HSA. GO-PEI affects HSA function by disrupting protein structure [27, 33]. The presence of p-GO causes changes in HSA and a defect in its binding capacity to bilirubin, leading to potential toxicity [27].
An interesting studied by Lee et al., is related to our study, and the results are remarkable. In this study, it was shown that BSA was broken down by GO, and the reaction rate increased with increasing temperature, incubation time and amount of GO [30]. To investigate the effect of GO on BSA hydrolysis, BSA was incubated with GO in phosphate buffer (pH=8.0) at different temperatures (4, 25, 40, 60 and 80℃) for 25 min. The BSA content of the supernatant after centrifugation decreased by 11%, 41%, 78% and 87% at 25, 40, 60 and 80℃, respectively, compared with the sample at 4℃. Because GO can form noncovalent protein complexes with protein, this study examined whether the decrease in BSA content in the supernatant could be due to noncovalent interactions between GO and BSA. The sediment from the centrifuge was analyzed by SDS-PAGE. The results show that the amount of BSA in sediment was the same in all samples, which indicates that the complete decrease in BSA is due to BSA cleavage and not BSA uptake on GO[30] .
A comparison of the effect of GO on HSA and BSA has been shown in a study in which GO can neutralize BSA fluorescence and slightly increase HSA fluorescence. In terms of fluorescence spectroscopy, the main difference between the two proteins is the number of Trp amino acids. BSA has two Trp residues (Trp 135 and Trp 214) and HSA has only one (Trp 214). Because Trp214 is in the same environment in both proteins, the quenching effect of GO is lower for BSA due to the presence of excess Trp [18].
Conclusion
Comparing the binding of albumin on the surface of graphene and GO, the results showed that although the binding energy of proteins to graphene is higher than that of GO. GO has a greater effect on the secondary structure. It is true that graphene and GO affect the microenvironment of the protein and reduce the amount of secondary structure to some extent, but in general the amount of change is dependent on the ratio of graphene or GO to albumin, temperature and pH. Depending on the above factors, the rate of structural and functional change in albumin may vary from unchanged to nonfunctional albumin.
Acknowledgment
This work was supported by Fund of Iran University of Medical Sciences (98-3-32-16262).
Conflict of Interest
The authors declare no conflicts of interest.
Reference
[1] Z. Behroozi, B. Rahimi, M.R. Hamblin, et al. Injection of cerium oxide nanoparticles to treat spinal cord injury in rats. Journal of Neuropathology & Experimental Neurology, 2022, 81: 635–642. https://doi.org/10.1093/jnen/nlac026
[2] Z. Sharifiaghdam, F. Dalouchi, M. Sharifiaghdam, et al. Curcumin-coated gold nanoparticles attenuate doxorubicin-induced cardiotoxicity via regulating apoptosis in a mouse model. Clinical and Experimental Pharmacology and Physiology, 2022, 49: 70–83. https://doi.org/10.1111/1440-1681.13579
[3] Z.S. Hoseini, A. Hajizade, A.J. Easton, et al. A meta-analysis of the efficiency of metal nanoparticles in vaccine delivery against infectious disease. Nanomedicine, 2021, 16: 481–495. https://doi.org/10.2217/nnm-2020-0358
[4] K. Zamani, N. Allah-Bakhshi, F. Akhavan, et al. Antibacterial effect of cerium oxide nanoparticle against Pseudomonas aeruginosa. BMC Biotechnology, 2021, 21: 68. https://doi.org/10.1186/s12896-021-00727-1
[5] A. Janzadeh, M.R. Hamblin, N. Janzadeh, et al. The toxic effect of silver nanoparticles on nerve cells: A systematic review and meta-analysis. Reviews of Environmental Contamination and Toxicology. Cham: Springer, 2021: 93–119. https://doi.org/10.1007/398_2021_67
[6] Janzadeh, Z. Behroozi, F. Saliminia, et al. Neurotoxicity of silver nanoparticles in the animal brain: A systematic review and meta-analysis. Forensic Toxicology, 2022, 40: 49–63. http://dx.doi.org/10.1007/s11419-021-00589-4
[7] Z. Behroozi, B. Rahimi, K. Kookli, et al. Distribution of gold nanoparticles into the brain: A systematic review and meta-analysis. Nanotoxicology, 2021, 15: 1059–1072. https://doi.org/10.1080/17435390.2021.1966116
[8] F. Ramezani, F. Nasirinezhad, N. Abotaleb. A review of nanotechnology strategies for neuron regeneration after spinal cord injury. Journal of Medical Physiology, 2016, 1(2): 42–54. http://jphysiology.com/index.php/jmp/article/view/9/8
[9] A.N. Banerjee. Graphene and its derivatives as biomedical materials: Future prospects and challenges. Interface Focus, 2018, 8: 20170056. https://doi.org/10.1098/rsfs.2017.0056
[10] P.V. Shinde, M. Saxena, M.K. Singh. Recent developments in graphene-based two-dimensional heterostructures for sensing applications. Fundamentals and Sensing Applications of 2D Materials. Amsterdam: Elsevier, 2019: 407–436. https://doi.org/10.1016/b978-0-08-102577-2.00011-7
[11] X.M. Li, L. Tao, Z.F. Chen, et al. Graphene and related two-dimensional materials: Structure-property relationships for electronics and optoelectronics. Applied Physics Reviews, 2017, 4: 021306. http://dx.doi.org/10.1063/1.4983646
[12] J.P. Nicholson, M.R. Wolmarans, G.R. Park. The role of albumin in critical illness. British Journal of Anaesthesia, 2000, 85: 599–610. https://doi.org/10.1093/bja/85.4.599
[13] T.R. Figueira, A.E. Vercesi, H.C.F. Oliveira. Lack of plasma albumin impairs intravascular lipolysis and explains the associated free fatty acids deficiency and hypertriglyceridemia. Lipids in Health and Disease, 2010, 9: 146. https://doi.org/10.1186/1476-511x-9-146
[14] D.G. Levitt, M.D. Levitt. Human serum albumin homeostasis: A new look at the roles of synthesis, catabolism, renal and gastrointestinal excretion, and the clinical value of serum albumin measurements. International Journal of General Medicine, 2016, 9: 229–255. https://doi.org/10.2147/ijgm.s102819
[15] S. Walayat, D. Martin, J. Patel, et al. Role of albumin in cirrhosis: From a hospitalist’s perspective. Journal of Community Hospital Internal Medicine Perspectives, 2017, 7: 8–14. https://doi.org/10.1080/20009666.2017.1302704
[16] F. Ramezani, H. Rafii-Tabar. An in-depth view of human serum albumin corona on gold nanoparticles. Molecular BioSystems, 2015, 11: 454–462. http://dx.doi.org/10.1039/C4MB00591K
[17] F. Ramezani, M. Amanlou, H. Rafii-Tabar. Gold nanoparticle shape effects on human serum albumin corona interface: A molecular dynamic study. Journal of Nanoparticle Research, 2014, 16: 2512. http://dx.doi.org/10.1007/s11051-014-2512-1
[18] S.H. Li, A.N. Aphale, I.G. Macwan, et al. Graphene oxide as a quencher for fluorescent assay of amino acids, peptides, and proteins. ACS Applied Materials & Interfaces, 2012, 4: 7069–7075. https://doi.org/10.1021/am302704a
[19] R. Feng, Y.P. Yu, C.X. Shen, et al. Impact of graphene oxide on the structure and function of important multiple blood components by a dose-dependent pattern. Journal of Biomedical Materials Research Part A, 2015, 103: 2006–2014. https://doi.org/10.1002/jbm.a.35341
[20] F. Ramezani. Effects of orientation on the stability and affinity of antibody-GNP conjugation. The Protein Journal, 2019, 38: 134–141. http://dx.doi.org/10.1007/s10930-019-09812-z
[21] S. Hashemzadeh, F. Ramezani, H. Rafii-Tabar. Study of molecular mechanism of the interaction between MEK1/2 and trametinib with docking and molecular dynamic simulation. Interdisciplinary Sciences: Computational Life Sciences, 2019, 11: 115–124. http://dx.doi.org/10.1007/s12539-018-0305-4
[22] F. Ramezani, M. Habibi, H. Rafii-Tabar, et al. Effect of peptide length on the conjugation to the gold nanoparticle surface: A molecular dynamic study. DARU Journal of Pharmaceutical Sciences, 2015, 23: 9. http://dx.doi.org/10.1186/s40199-014-0085-2
[23] M. Vahed, F. Ramezani, V. Tafakori, et al. Molecular dynamics simulation and experimental study of the surface-display of SPA protein via Lpp-OmpA system for screening of IgG. AMB Express, 2020, 10: 161. http://dx.doi.org/10.1186/s13568-020-01097-1
[24] M. Abbasi, F. Ramezani, M. Elyasi, et al. A study on quantitative structure-activity relationship and molecular docking of metalloproteinase inhibitors based on L-tyrosine scaffold. DARU Journal of Pharmaceutical Sciences, 2015, 23: 29. http://dx.doi.org/10.1186/s40199-015-0111-z
[25] X.X. Ba, T. Gao, M. Yang, et al. Thermodynamics of the interaction between graphene quantum dots with human serum albumin and γ-globulins. Journal of Solution Chemistry, 2020, 49: 100–116. http://dx.doi.org/10.1007/s10953-019-00941-8
[26] Z.J. Ding, H.W. Ma, Y.Y. Chen. Interaction of graphene oxide with human serum albumin and its mechanism. RSC Advances, 2014, 4: 55290–55295. http://dx.doi.org/10.1039/C4RA09613D
[27] Z.J. Ding, Z.J. Zhang, H.W. Ma, et al. In vitro hemocompatibility and toxic mechanism of graphene oxide on human peripheral blood T lymphocytes and serum albumin. ACS Applied Materials & Interfaces, 2014, 6: 19797–19807. https://doi.org/10.1021/am505084s
[28] S. Huang, H. Qiu, S. Lu, et al. Study on the molecular interaction of graphene quantum dots with human serum albumin: Combined spectroscopic and electrochemical approaches. Journal of Hazardous Materials, 2015, 285: 18–26. http://dx.doi.org/10.1016/j.jhazmat.2014.11.019
[29] A. Khan, F. Khan, M. Shahwan, et al. Mechanistic insight into the binding of graphene oxide with human serum albumin: Multispectroscopic and molecular docking approach. Spectrochim. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2021, 256: 119750. https://doi.org/10.1016/j.saa.2021.119750
[30] H. Lee, M.H. Tran, H.K. Jeong, et al. Nonspecific cleavage of proteins using graphene oxide. Analytical Biochemistry, 2014, 451: 31–34. https://doi.org/10.1016/j.ab.2014.01.017
[31] X. Liu, C. Yan, K.L. Chen. Adsorption of human serum albumin on graphene oxide: Implications for protein Corona formation and conformation. Environmental Science & Technology, 2019, 53: 8631–8639. https://doi.org/10.1021/acs.est.8b03451
[32] Z.Z. Nan, C.C. Hao, X.Q. Ye, et al. Interaction of graphene oxide with bovine serum albumin: A fluorescence quenching study. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy, 2019, 210: 348–354. https://doi.org/10.1016/j.saa.2018.11.028
[33] Šimšíková M. Interaction of graphene oxide with albumins: Effect of size, pH, and temperature. Archives of Biochemistry and Biophysics, 2016, 593: 69–79. http://dx.doi.org/10.1016/j.abb.2016.02.015
[34] B.B. Sun, Y.Q. Zhang, W. Chen, et al. Concentration dependent effects of bovine serum albumin on graphene oxide colloidal stability in aquatic environment. Environmental Science & Technology, 2018, 52: 7212–7219. https://doi.org/10.1021/acs.est.7b06218
[35] Y. Sun, J.L. Ding, H. Zong, et al. Molecular dynamics simulations of albumin and graphene structures and their interaction. Journal of Southeast University (Natural Science Edition), 2014, 44: 123–128. http://dx.chinadoi.cn/10.3969/j.issn.1001-0505.2014.01.022
[36] P. Hampitak, D. Melendrez, M. Iliut, et al. Protein interactions and conformations on graphene-based materials mapped using a quartz-crystal microbalance with dissipation monitoring (QCM-D). Carbon, 2020, 165: 317–327. https://doi.org/10.1016/j.carbon.2020.04.093
Copyright© Hossein Arzani and Fatemeh Ramezani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.