Research Article
NRAMP1 Gene Polymorphism and Susceptibility to Cutaneous Leishmaniasis in Iraq
Ghada Basil Alomashi, Hasan Raheem Khudhur *
College of Medicine, University of Al-Qadisiyah, Iraq.
* Corresponding author. E-mail: hassanraheem1990@gmail.com
Received: Nov. 1, 2017; Accepted: Dec. 25, 2017; Published: Dec. 29, 2017
Citation: Ghada Basil Alomashi, and Hasan Raheem Khudhur, NRAMP1 Gene Polymorphism and Susceptibility to Cutaneous Leishmaniasis in Iraq. Nano Biomed. Eng., 2017, 9(4): 333-336.
DOI: 10.5101/nbe.v9i4.p333-336.
Abstract
Cutaneous leishmaniasis (CL) is a vector-borne disease and endemic in most regions of Iraq, especially in the regions with poor populations. Natural resistance associated macrophage protein 1 (NRAMP1) gene plays an essential role in susceptibility to CL and disease pathology. This study aimed to study the polymorphism in NRAMP1 gene, and tried to identify an association between gene variants and susceptibility to CL infection in Iraqi population / AL-Muthanna province. Samples of peripheral blood were collected from 60 patients with CL and 32 apparently healthy controls. NRAMP1 (D543N) polymorphism was detected in patients and control groups by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique. The results indicated a statistically significant difference in genotype distribution between CL cases and healthy controls (p = 0.036), and the results indicated that genetic variations of D543N were not associated with susceptibility to CL infection, and the frequency of allele A was greater in controls than in patients with statistical significance of p = 0.01.
Keywords: Cutaneous leishmaniasis; NRAMP1 polymorphisms; D543N; Iraq
Introduction
Cutaneous leishmaniasis (CL), a vector-borne disease caused by obligate intra-macrophage protozoa, transmitted by the species of phlebotomine sandflies, and characterized by diversity and complexity [1]. CL is a very old disease in Iraq, also called "Baghdad boil" and caused by L. major and L. tropica [2]. CL is the most common form of leishmaniasis, with about 1.5 million cases in Iraq every year, and about 50 to 70% of all cases in the world [3]. Natural resistance-associated macrophage protein 1 (NRAMP1) gene is an important member of the solute carrier family 11 and called SLC11A1 [4]. In humans, NRAMP1 gene is located in the chromosome region 2q35, with 16 exons [5]. In a resting macrophage, the protein encoded by NRAMP1 gene is assembled into the membrane of late endosome, while the following phagocytosis is relocated to the membrane of phagosome [6, 7]. NRAMP1 protein transfers divalent metal ions across the phagosomal membrane and might be a critical factor for resistance to some microbial infections, NRAMP1 influences a variety of antimicrobial responses of a macrophage, including induction of radical oxygen and nitric oxide intermediates, production and activation of various pro-inflammatory cytokines (TNF-α and IL-1β) and regulation of anti-inflammatory cytokine IL-10 [8, 9]. Due to the importance of NRAMP1, especially D543N (rs17235409) polymorphisms in the control of intracellular infections [10], the associations of the mentioned polymorphisms with susceptibility to CL infection were investigated in this study.
Experimental
Subjects and study design
A total of 60 cases of suspected CL were included in this study. Their mean age was 17 ± 13 years old, including 34 male and 26 female. The patients complained of a skin lesion in an exposed part of the body mostly in the face, leg and arm. Cases diagnosed clinically by a special dermatologist as CL and confirmed as CL patients based on clinical symptoms and parasitological parameters [11]. The control group included 32 apparently healthy individuals; their mean age was 21 ± 9 years old, including 18 males and 14 females.
D543N (rs17235409) typing
Genomic DNA from blood samples was extracted by using Geneaid DNA Extraction Kit (Whole Blood), according to the manufacturers’ instructions. Genotyping for D543N polymorphisms was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) with two primers as described by Fattahi-Dolatabadi et al. [10], and provided by Bioner Company as listed in Table 1. The volume of PCR mixture for polymorphisms was 50 μl, including 1× premaster mix (Bioneer, Korea), 2.5 L of each primer (10 pm/L), 5 μL of genomic DNA (20 ng/L), and the volume was completed with nuclease-free water. Thermocycler conditions were: 94 °C for 4 min, and then 38 cycles of 94 °C for 60 sec, 58 °C for 90 sec, and 72 °C for 40 sec followed by 10-min extension at 72 °C. Then products of PCR were analyzed by electrophoresis in a 1% agarose gel and stained with 10 mg/mL ethidium bromide. The PCR products were digested by using restriction enzyme Avall. Then PCR-RFLP master mix was carried out according to instructions of the company Biolabs, U.K. After that, PCR-RFLP product was analyzed by electrophoresis of 2.5 % agarose gel.
Table 1 The sequence of primers, restriction enzyme and DNA fragments size
|
Name |
Primer sequence |
R. Enzyme |
Genotype |
Products |
|
D543N |
F:5’-ACT-AAGAAA-GAC-CCG-AGG-C-3 |
Avall |
GG |
126,79 and 39 bp |
|
R:5’-GGG-GCA-CGT-TGG-TGTTTA-C-3 |
GA |
126,79,39 and 205 bp |
||
|
AA |
205 and 39 bp |
Statistical analysis
The result was analyzed by using the statistical software package SPSS 23. The frequency of allele and genotyping of D543N were performed for each polymorphism among patient and control groups by using χ2-test and odds ratio with 95% confidence [12]. The probability of p ≤ 0.05) was considered statistically significant.
Results and Discussion
In total, 60 CL cases and 32 controls were genotyped; equilibrium of Hardy-Weinberg was used in both cases and controls. Fig. 1 and Table 2 show the distribution of D543N genotypes and alleles among cases and controls that were detected by PCR-RFLP technique. At this locus were three genotypes: GG, GA and AA with band size 126/79/39 pb, 205/ 126/79/39 pb and 205/39 pb, respectively. A significant difference in genotype distribution was recorded between CL cases and healthy controls (p = 0.036). Also, there was a significant difference found in G and A allele between patients and healthy controls; however, data in the present study revealed higher frequency of allele A in controls than in the CL patients, as of 31.25% and 15%, respectively (p = 0.01). Genetic factors play a crucial role in host susceptibility or resistance to infectious diseases; variations within the NRAMP1 gene were assessed in this study with respect to CL in Al-Muthama province, Iraq which has not been reported before from this region. The current study showed that effects of D543N polymorphisms on susceptibility to CL, and the difference in frequency of alleles and genotypes D543N between patients and controls was statistically significant (p = 0.036). The current data finding was in agreement with Fattahi-Dolatabadi et al. [10]. At the same time, there were some exceptions such as reported by Ortiz-Flores et al. and Sophie1 et al. [13, 14], which showed no significant difference of alleles between CL patients and healthy control. The present study revealed no association between D543N polymorphisms and CL, and the frequency of allele A was greater in controls than in the patients (p = 0.01). The results were in agreement with the report by Samaranayake et al. [15] in Sri Lanka which showed a lack of association between the GA genotype and CL infection, and also consistent with the findings by Sophie et al. [14] in Pakistan, which showed there was no association between D543N polymorphisms and CL. On the another hand, Fattahi-Dolatabadi et al. [10] in Iran reported an association between the GA genotype and CL infection. However, other factors should be taken into account such as that infecting Leishmania species are not the same in all cases. There is a high percentage of Leishmania as the major reason for most infections in the areas of Iraq, as well as different vector and animal host species found in areas of Iraq [2]. In addition, host’s genetic factors play an essential role in host susceptibility, and the possibility of more SNPs than D543N should be included to explain the inefficiency of NRAMP1.
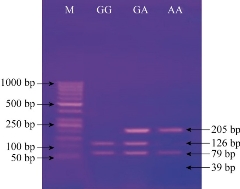
Fig. 1 Agarose gel electrophoresis image that shows the PCR-RFLP product analysis of NRAMP1 (D543N) gene by using AvaII restriction enzyme. M: marker (1000-50 bp). Lane GG homozygote: The product was digested by a restriction enzyme at 126 and 79 bp, and invisible at 39 bp bands. Lane G/A heterozygote: The product was digested by restriction enzyme into 126, 79 and 205 bp, and invisible at 39 bp bands. Lane AA homozygote: The product was digested by restriction enzyme into 205 bp and invisible at 39 bp bands.
Table 2 Distribution of NRAMP1 (D543N) genotypes and alleles
|
Variable |
Patients (60) |
Controls (32) |
P value |
Odd Ratio |
95% Confidence Interval |
Overall P |
|
|
Lower |
Lower |
||||||
|
GG |
44 (73.30%) |
18 (56.25%) |
0.096 |
2.139 |
0.867 |
5.276 |
0.036* |
|
GA |
14 (23.3%) |
8 (25%) |
0.858 |
0.913 |
0.336 |
2.480 |
|
|
AA |
2 (3.3%) |
6 (18.75%) |
0.012* |
0.149 |
0.028 |
0.790 |
|
|
Allele |
|
|
|
|
|
|
|
|
G |
102 (85%) |
44 (68.75%) |
0.01* |
2.576 |
1.243 |
5.336 |
0.01*
|
|
A |
18 (15%) |
20 (31.25%) |
0.01* |
0.388 |
0.187 |
0.804 |
|
* represents a significant difference at 0.05 level.
Conclusions
The results of the present study indicated that the genetic variations in NRAMP1 (D543N) gene were not associated with the susceptibility to CL infection in the sample population from Iraq.
Conflict of Interests
The authors declare that no competing interest exists.
References
Copyright© Ghada Basil Alomashi, and Hasan Raheem Khudhur. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.