Review
Biological Application of Digital Microfluidics Technology
Bin Liu1, Yan Deng1,2, Binbing Qin1, Zhiyang Li1, Nongyue He1,2 *
1State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, P.R. China;
2Hunan Key Laboratory of Green Packaging and Application of Biological Nanotechnology, Hunan University of Technology, Zhuzhou,412008, China
*Corresponding authors. Email: nyhe1958@163.com; nyhe@seu.edu.cn
Citation: B. Liu et al. Biological Application of Digital Microfluidics Technology. Nano Biomed Eng. 2010, 2(2), 149-154.
DOI: 10.5101/nbe.v2i2.p149-154
Abstract
Digital microfluidics technology offers a platform for developing diagnostic applications with the advantages of portability, sample and reagent volume reduction, faster analysis, increased automation, low power consumption, compatibility with mass manufacturing and high throughput. In addition to diagnostics, digital microfluidics is finding use in nucleic acid analysis, peptide and protein analysis, cell analysis, drug analysis and delivery and immunization analysis. In this review, we describe these applications, their implementation, and associated design issues. As other review in the digital microfluidics technology, there have been and will be unexpected developments as DMF matures, but we predict that the future is bright for this promising technology at the last section.
Keywords: Digital microfluidics technology; Nucleic acid analysis; Peptide and protein analysis; Cell analysis; Drug analysis and delivery
1. The Advent and Development of Digital Microfluidics Technology
Traditional (continuous-flow) microfluidic technologies are based on the continuous flow of liquid through microfabricated channels [1]. Continuous flow systems are inherently difficult to integrate because the parameters that govern flow field (e.g. pressure, fluid resistance, electric field strength) vary along the flow-path, making the flow at any location dependent upon the properties of the entire system.The concept of digital microfluidics (DMF) arose in the late 1990s and involves the manipulation of discrete volumes of liquids on a surface. Manipulation of drop- lets can occur through various mechanisms, including electrowetting [2], dielectrophoresis [3], thermocapillary transport [4] and surface acoustic wave transport [5].DMF was popularized in the early 2000s by Fair and coworkers [6] and Kim and coworkers [7] at Duke and UCLA, respectively. The technique was explained as being a phenomenon driven by surface tension, and was called “electrowetting” or “electrowetting on dielectric” (EWOD). A detailed review of electrowetting basics can be found in the work of Mugele [8]. In addition, work on simulation and modeling of droplet- based electrowetting has been reported by Biddut Bhat-tacharjee and Homayoun Najjaran [9]. In contrast to continuous flow biochips, digital microfluidic biochips platform is under software-driven electronic control, eliminating the need for mechanical tubes, pumps, and valves. Moreover, because each droplet can be controlled independently, these “digital” systems also have dynamic reconfigurability, whereby groups of cells in a microfluidic array can be reconfigured to change their functionality during the concur of the digital microfluidic platform, when considered in light of real applications, are as follows [10]:
(1) No moving parts. All operations take place between the two plates under direct electrical control without use of pumps or valves.
(2) No channels are required. The gap is simply filled with liquid; channels exist only in a virtual sense and can be instantly reconfigured through software.
(3) Many droplets can be independently controlled be- cause the electrowetting force is localized at the surface.
(4) Evaporation can be controlled or prevented with the oil surrounding the droplets.
(5) No ohmic current exists. Although capacitive cur- rents exist, the device blocks direct current, minimizing sample heating and electrochemical reactions.
(6) Works with a wide variety of liquids. Most electrolyte solutions will work.
(7)Near 100% utilization of sample or reagent is possible by wasting no fluid for priming channels or filling
reservoirs.
(8) Compatible with microscopy. Glass substrates and transparent indium-tin-oxide (ITO) electrodes make the chip compatible with observation from a microscope.
(9) Extremely energy efficient. Using nanowatts to microwatts of power per transfer.
(10) High speed. Droplet speeds of up to about 25 cm/s achieved.
(11) Droplet-based protocols functionally are equivalent to bench-scale wet chemistry. Thus, user can simple scale down, automate, and integrate established assays and protocols.
(12) Conditional execution steps can be implemented by direct computer.
2. Application of Digital Microfluidics Technology in Bioanalysis
To accomplish a digital microfluidics device it requires a hierarchical taxonomy. At the top level, applications are scaled to a microfluidic platform. The second level describes the microfluidic operations. And the third level describes the components to perform the operations. This manageable design approach is shown in Fig. 1. The top level in Fig. 1 includes biomedical applications such as electrophoretic separations, nucleic acid analysis, protein/enzyme analysis, immunoassays and bioassays, and pathogen detection. In the section below, these main applications will be discussed in detail. The second level of Fig. 1 decomposes the set of applications into common fluidic functions, such as liquid transport, mixing, filtering, and analysis. These common operations determine the requirements for the set of microfluidic hardware components in the third level, such as buffers, channels, reservoirs and mixers.
2.1 Nucleic acid analysis
Microfluidic technology has been successfully aplied for numerous nucleic acid applications. One promising application is for DNA amplification by using the polymerase chain reaction (PCR). Recently, Daxiang Cui et al reported a new PCR system based on mercaptoacetic acid-modified CdTe nanocrystals (mQDs) and can improve the sensitivity and specificity of PCR within less than 1.33 mg/mL [12], but compared with the conventional PCR assays, the concept of miniaturizing and automating PCR systems through digital microfluidics and advanced microfabrication techniques has attracted a great deal of attention because of the potential to dramatically improve the speed, portability, cost, sensitivity and specificity. For example, Zhishan Hua et al successfully developed a multiplexed realtime PCR system using electrowetting-based digital microfluidics [13]. The reproducibility and sensitivity of the digital microfluidic PCR system presented here compared favorably to conventional bench top real time PCR instruments but provided many advantages in terms of automation, cost and time to result. Yung, Tony K. F. et al developed a digital PCR based method for the quantitative detection of the two common epidermal growth factor receptor muta- tions in the plasma and tumor tissues of patients suffer- ing from non-small cell lung cancers [14]. The sensi- tive detection and accurate quantification of low abundance EGFR mutations in tumor tissues and plasma by microfluidics digital PCR would be useful for predicting treatment response, monitoring disease progression and early detection of treatment failure associated with acquired drug resistance. Lun FMF et al focused on comparing the imprecision of microfluidics digital PCR with that of a well-established non digital PCR assay for measuring male fetal DNA in maternal plasma [15]. They proved that microfluidics digital PCR represents an improvement over previous methods for quantifying fetal DNA in maternal plasma, enabling diagnostic and research applications requiring precise quantification. They predicted that this approach may also impact oth- er diagnostic applications of plasma nucleic acids, e.g., in oncology and transplantation. Another promising application is for low-cost and rapid-prototyping. Mohamed Abdelgawad et al developed new rapid prototyping techniques for digital mi- crofluidics using microcontact printing and laser printing on printed circuit board substrates [16]. Mohamed Abdelgawad et al also report two rapid prototyping techniques for the fabrication of DMF chips, making use of commercially available printed circuit board (PCB) substrates [17]. In the first method, actuation electrodes are patterned on PCB substrates by using photolithography, in a manner similar to what has been reported for other applications. This method is fast, inexpensive, and easy relative to conventional microfabrication. In the second technique, actuation electrodes are patterned directly onto substrates using a desktop laser printer; this method enabled ultra-high throughput fabrication.
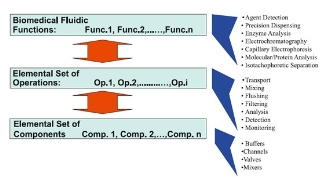
Figure 1. Hierarchical design approach for implement- ing numerous applications on a common set of compo- nents [11].
2.2 Peptide and protein analysis
Recently, the droplet-based (digital) microfluidics platform has been developed to prepare and purify protein samples for measurement by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS). Chatterjee D et al showed that a complete integrated sequence of protein processing steps could be per- formed on droplet-based microfluidics platform, in- cluding disulfide reduction, alkylation, and enzymatic digestion, followed by cocrystallization with a MALDI matrix and analysis of the sample in situ by MALDI- MS [18]. In 2009, Jebrail M J et al reported the devel- opment of an automated microfluidic method for ex- tracting proteins from heterogeneous fluids by precipitation [19]. As showed in Fig. 2, the structure of device consists of the four reagent reservoirs, the waste reser- voir, and extraction electrode. Frames from a movie depicting the extraction and purification of BSA (50 mg/mL) in 20 % TCA (precipitant) and washing with 70/30 v/v chloroform/acetonitrile (rinse solution). In the final frame, the precipitated protein is redissolved in a droplet of 100 mM borate buffer containing 1% SDS. It is the first microfluidic method for extracting proteins from heterogeneous fluids by precipitation. The digital microfluidics used in the method not only facilitates high throughput extraction and screening of proteins, but also has reduced analysis time. Experiment results suggested great potential for the development of integrated, multi-step processes incorporating sample reduction, alkylation and digestion. Especially this work represented an important first step in our efforts to develop fully automated microfluidic methods for proteomic analyses.
2.3 Cell analysis
There are several new techniques to analyze the cell, such as new capillary electrophoresis, molecular imaging [20, 21], single walled carbon nanotubes [22] and microfluidic chip. Conventional flow cytometers which are routinely utilized for analyzing the physical and chemical properties of biological cells require a high amount of reagent for analysis and furthermore are both bulky and expensive and require trained personnel for operation and maintenance. In 2008, Barbulovic Nad I et al introduced a new method for implementing cell-based assays [23]. This method is based on digital microfluidics which is used to actuate nanolitre drop- lets of reagents and cells on a planar array of electrodes. Experiments demonstrated that this method is advantageous for cell-based assays because of automated manipulation of multiple reagents in addition to reduced reagent and analysis time. In 2010, Jiajia Ji et al re- views advances of nanotechnology in the stem cells research, including various micro/nanofabrication technologies, microgrooves technology and so on [24]. To circumvent the complexity of the detection systems of microfluidic devices, Hartley et al recently reported on a CMOS optical active pixel sensor (APS) for near-field detection and counting of microscopic particles [25]. To further enhance the digital cytometric capabilities of the original sensor, Yahya Hosseini et al modified and utilized a dual APS-array scheme to faci- litate the determination of the velocity and size of par- ticles flowing in microfluidic channels [26]. The experimental results suggested that the dual-array-APS arrangement offered a simplistic yet effective strategy for size measurements of microscopic particles in micro- fluidic environments.
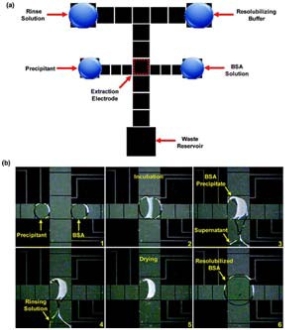
Figure 2. Digital microfluidic device and method for protein precipitation [19] .
2.4 Drug analysis and delivery
Recent advances in molecular biology and genetic research have made possible the creation of more powerful and effective cancer therapeutics, bringing about the realization of the century old concept of ‘‘magic bullets’’ that can carry therapeutic drugs to target sites with high specificity [27]. In 2009, Kanaka Hettiarachchi and Abraham P. Lee reported the ability to generate functionalized multilayer gas lipospheres with precisely controlled size and drug carrying capaci ty [28]. Through their previous work, they demonstrat- ed that the application of multilayer lipospheres as drug delivery agents, although those vehicles are produced with techniques that resulted in size and loading inconsistency. Tatiana N Laremore et al reviewed the recent progress and applications in glycosaminoglycan and heparin research [29]. They showed that digital microfluidics may provide environments that more accurately mimic biological synthesis and can be manipulated leading to new biosynthetic and biological screening designs. They predicted that the future should bring an improved understanding of Golgi-based heparin/HS biosynthesis, its control and scale-up, and the rapid, microscale analysis of heparin/HS leading to the pro- duction of bioengineered anticoagulant heparin and a novel array of designer heparin-based therapeutics.
2.5 Immunization analysis
Immunoassays and enzyme activity assays currently used in newborn screening have been translated to a disposable microchip programmed to dispense, trans- port, mix, wash, and incubate individual microdroplets from specimens, including dried blood spot extracts, and reagents all under software control. There are many characteristics which make digital microfluidic technology attractive to newborn screening, such as low volumes, direct translation of existing assays, automation, portability, inexpensive manufacturing, sam- ple compatibility, scalability and multifunctionality. David S. Millington et al described a cost-effective new platform that reduced the time to result reporting and could perform multiplexing assays requiring different platforms [30]. Chun-Che Lin et al have a brief introduction to microfluidic immunoassays and showed the critical issues which are important for microfluidic immunoassays and should be addressed properly [31]. The critical issues are as follows:
(1) Mass Production. For commercial and disposability considerations, Materials with similar properties and capabilities for mass production by techniques, such as injection molding and embossing are in great demand for practical applications.
(2) Multiplicity. The multiplexed assay will continue to be the dominant method for commercialization of these microfluidic immunoassays.
(3) Surface Modification and Immobilization. Nonspecific adsorption or binding to molecules rather than analytes is a key concern in immunoassays because it may greatly impair the sensitivity and selectivity.
(4) Purification and Concentration. Due to the complexity and the low trace of analytes in biosamples, purification and concentration steps are often required in microfluidic immunoassays.
(5) Detection. Developing and integrating miniaturized, compact, portable, and inexpensive detection systems with an acceptable sensitivity onto microfluidic devices are still in great demand.
(6) Integration, Packaging, and Cost down issues. The ultimate goal for practical commercialization is to develop fully integrated, well packaged, disposable, and cheap microfluidic systems for immunoassays.
(7) Storage of Reagents. This is one important issue in portable devices because most bioreagents are not durable under room temperature and some of them require special environments for storage.
3. The Prospect of Digital Microfluidics Technology
Like many new technologies, DMF has obviously shortcuts in the previous years. Most digital microflui- dic platforms built to date are highly specialized and designed to fulfill the requirements of a single particular application within a limited set of operations. The detection techniques should be able to circumvent the limitations of hydrophobic surfaces and exploit the ad- vantages of the array format, high droplet transport speeds and rapid mixing schemes. As the applications discussed in this article, we pro- pose that the ever-expanding community of DMF re- searchers (including academics, ALL, and others) will solve some of the mechanistic and practical problems that remain, such that DMF will become a widely prac- ticed technique in the next decade.
Acknowledgments
This research is financially supported by the National Natural Science Foundation of China (NO. 60927001, 60971045), the National Key Program for Developing Basic Research (2010CB933903, 2007CB936104), the Chinese 863 High Tech Project (NO.2007AA022007), and China National Science and Technology Major Projects (2009ZX10004-311).
ReferenceS
Received 15 May, 2010; accepted 22 June, 2010; published online 18 July, 2010.
Copyright: (c) 2010 B. Liu et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.