Article
Hepatocarcinoma Single Cell Migration on Micropatterned PDMS Substrates
Bin Zheng1, Shuling Hsieh2, Chihchung Wu3, Chunhsin Wu4, Peiying Lin5, Chiungwen Hsieh5, Itin Li5, Yunshan Huang6 , Huaymin Wang7 and Shuchen Hsieh5*
1School of Materials Science and Engineering, Dalian University of Technology, Dalian, Liaoning 116024, People’s Republic of China
2Department of Seafood Science, National Kaohsiung Marine University, Kaohsiung 81157, Taiwan
3Department of Nutrition and Health Sciences, Chang Jung Christian University, Tainan, 71101 Taiwan
4Department of Computer Science and Information Engineering, National University of Kaohsiung, Kaohsiung, Taiwan 5Department of Chemistry, Center for Nanoscience and Nanotechnology, National Sun Yat-sen University Kaohsiung, Taiwan 6Department of Food Science and Technology, Tajen University, Pingtung, 90741,Taiwan
7Kaohsiung Veterans General Hospital, Kaohsiung, 81362, Taiwan
* Corresponding author: shsieh@facmail.nsysu.edu.tw (Shuchen Hsieh)
Cell migration influences many normal and pathological processes and is one of key issues addressed in cancer research studies. In this report, a plasma patterned polydimethylsiloxane (PDMS) substrate was used to selectively position hepatocarcinoma cells in order to characterize their migration behavior. We observed that cell mobility was directly related to the differentiation stage of the cells, with poorly-differentiated (SK-Hep-1) cells exhibiting higher mobility that well-differentiated (Hep-G2) cells. We propose that this difference occurs due to a loss of adhesion molecules presented at the apical membranes of the poorly-differentiated SK-Hep-1 cells, thereby reducing their adhesion to the surface. Our results provide new insight into the relationship between carcinoma cell differentiation grade and mobility. Further this experimental process may provide a simple and effective model for universal cell biology studies and applications in microsystems technology.
Keywords: Cell adhesion, Cell spreading, Hepatocyte, Micropatterning, Polydimethylsiloxane
Citation: B. Zheng, et al. Hepatocarcinoma single cell migration on micropatterned PDMS substrates. Nano Biomed. Eng. 2011, 3(2), 99-106.
DOI: 10.5101/nbe.v3i2.p99-106.
Hepatocellular carcinoma (HCC), which is known to be involved in chronic hepatitis and progressive liver cirrhosis, has a high mortality rate even today [1]. It is known that cell migration plays a key role in tumor invasion and metastasis, and a variety of other normal and pathological processes throughout the cycle of life, from embryonic development to death, including birth defects, developmental morphogenesis, angiogenesis, wound healing, and tissue repair [2, 3]. Therefore, understanding the principles of cell migration and its related influence is expected to provide insight into HCC progression, and may lead to new strategies for disruption, prevention, or cure.Cell migration is complex, consisting of multiple simultaneous dynamic processes. For example, surface attachment and detachment cycles, development and collapse of filopodia, movement of the cell body center, and maintenance of cell morphology [4], as well as environmental cues, such as interaction with the extracellular matrix (ECM), chemicals present in theenvironment, and other cells [2, 5, 6], all contribute to this dynamic process. Simply put, cell migration arises from spontaneous movement as well as tactile response to environmental signals, each playing essential roles in various cellular functions [2, 4, 7]. In the past few years, great strides have been made in understanding and predicting the two types of cell migration in population- based systems, however, to our knowledge, studies of single-cell migration, which are even more critical to elucidate the fundamental causes of spontaneous movement at the molecular level, are scarce [3, 4, 6]. One of the primary reasons for this may be the lack of proper single-cell assays, which can disperse cells and be used to analyze single-cell activity as a function of time [8]. Therefore, the quest for a single-cell based system and for building a non-continuum movement model is urgent. Cell micropatterning is a good candidate for studying single-cell movement, as it provides a mechanism for dynamic modulation of the interaction between cells and materials at the molecular level [6], and has been successfully used in biosensor fabrication, tissue engineering, bioelectronics, and basic biology studies [9- 14].In this study, we have used patterned poly-dimethylsiloxane (PDMS) substrates to position and then monitor the migration behavior of well and poorly-differentiated HCC cells. PDMS substrates are biocompatible, durable, transparent, and flexible and have therefore been used frequently in cell patterning research [15-19]. However, PDMS is naturally hydrophobic, and thus it is difficult to attach some types of cells on this low surface energy substrate [20]. Researchers have used different adhesion promoters on PDMS surfaces to enhance cell attachment, including hydrophobin [9], Laminin [16], poly L-Lysine [21], vironectin [22], and self-assembled monolayers [23]. Also, plasma patterning has been used to modify PDMS surfaces to create patterned oxidized regions that exhibit hydrophilic character and can be used for cell patterning [24]. In our published work in this area, plasma patterned PDMS surfaces were used to fix ferritin proteins [25]. In this study, plasma patterned PDMS surfaces were used as an adherent substrate to provide a model system for studying the cellular motion of HCC cells with different histological grades.
PDMS (Sylgard 184, Dow Corning, Midland, MI, USA) was composed of a 10:1 (w : w) ratio of pre- polymer to curing agent, which was thoroughly mixed, allowed to cure for 5 min, and then poured into a steel washer that was placed on a clean glass microscope slide. The resulting PDMS samples (1–2 mm thick) were then cured in an oven for 2 h at 70 °C. In order to prevent air bubbles from forming in the PDMS samples, the solidification process required heating in a closed vessel. A copper TEM grid (400 mesh EMITECH – square type – 3.2 mm diameter) was placed on the cured PDMS surfaces and then the samples were exposed to a low- pressure “air” plasma (53.3 Pa, 2 min, 10.5 W – Harrick Scientific Products, Inc. Model PDC-001) to selectively generate oxidation patterns on the hydrophobic PDMS surfaces. Details of the substrate preparation and micropatterning method are described elsewhere [26].
Human Hep-G2 and SK-Hep-1 cell lines were purchased from Bioresource collection and research center, HsinChu, Taiwan. They were cultured in Du l b e c c o 's Mo d i f i e d E a g l e Me d i u m ( DME M) supplemented with 10% fetal bovine serum (FBS) (Gibco, Invitrogen, Carlsbad, California) at 37 ºC in a humidified atmosphere of 5% CO2. Before patterning, the cells were rinsed with 5 mL phosphate buffer solution (PBS pH 7.4), treated with 1 mL Trypsin–EDTA (0.25%) and swayed for 5 min to insure thorough mixing. The cell suspension was then washed into a sterile centrifuge tube using 5 mL of PBS medium. After being centrifuged for 2 min at 1000 rpm, the cells were re-suspended in medium and the supernatant removed. Media was added to the resulting cells which were distributed into 24 prepared well plates for culture in an incubator at 37 °C and a humidified atmosphere of 5% CO2 and 95% air.
MTT dye solution was taken up by the cells through direct interaction with dehydrogenase enzymes in the living cell’s mitochondria. The MTT was reduced to purple formazan which formed crystals that accumulated within the living cells. Solubilization of the cells resulted in the liberation of the purple product, which was detected using colorimetric measurement. The ability of cells to reduce MTT provides an indication of the mitochondrial integrity and activity which, in turn, may be interpreted as a measure of the cell number/proliferation/viability/ survival/toxicity. For viability testing, MTT cell proliferation assays were carried out. First, an 8×102 cells mL-1 suspension was seeded in 24-well plate patterned PDMS samples and cultured for 24 h in an incubator at 37°C in a 5% CO2 atmosphere. Once adhesion was verified, residual media was washed away using PBS. Cells on every well plate were incubated with 50 μL 0.1% MTT and 450 μL medium for an additional 3 h. After cell treatment, absorbance at 570 nm was measured with a microplate reader.
Optical images of the cell samples were acquired using an inverted optical microscope (A1-Mat, Carl Zeiss, Jena, Germany) with a CCD camera. Images of several different regions of each sample surface were collected at regular intervals starting from cell seeding, to monitor changes in cellular position and organization over time.
Motivation for studying cell movement at single cell levels in vitro is that by concentrating cells into small, precisely and regularly sized and shaped areas, these “seeds” can act as starting points for cellular migration. In general, oxidized, charged, and hydrophilic surfaces have been used extensively for cell adhesion and growth. For the native hydrophobic surfaces of cover glass and PDMS, cell adhesion promoters are routinely deposited prior to cell seeding to aid cell adherence and growth [9, 16, 21-23]. However, the introduction of these adhesion promoters may affect cell migration through chemical interactions, thus influencing the migration behavior. As discussed above, plasma treatment is commonly used to oxidize hydrophobic polymer surfaces, transforming the surface free energy to a hydrophilic state [24, 26]. Our plasma-based method allows us to further control the chemical pattern (size and shape) through selective oxidation of hydrophobic PDMS to create samples for cell patterning that do not require substrate adhesion molecules. We compared HCC cell attachment on cover glass substrates with that on plasma treated PDMS, as shown in Fig. 1. Cells on the cover glass were randomly distributed and tended to aggregate, while, cells on the patterned PDMS substrates attached preferentially according to the patterned regions of the surface. Further, the SK-Hep-1 cells appeared to be more strongly influenced by the substrate pattern than the Hep-G2 cells.Cell viability tests were performed using the Hep-G2 and SK-Hep-1 cells on plasma patterned PDMS substrates to insure that they were sufficiently biocompatible for time course migration studies. MTT assay results from viability testing are shown in Fig. 2. Three separate assay tests were performed on the Hep-G2 and SK-Hep-1 cells on PDMS samples and the average values for cell viability were 74% and 70%, respectively. Literature reports have shown the use of untreated PDMS surfaces results in high levels of cell death (45% and 93.4% for 3T3 and bovine aortic endothelial cells, respectively), however, acetone plasma treatment of PDMS surfaces showed significant improvement [27]. The plasma-patterned PDMS surfacesused in this study to position HCC cells exhibited much lower toxicity (higher cell viability values) which is consistent with the reported results. Although the cell viability on PDMS surface is less than 100% (as defined by cell viability on a cover glass), it is nonetheless high enough for the purpose of migration studies.Cell activity is controlled by chemical and mechanical interactions between cells, and between cells and their surrounding environment. In vitro simulation of the cell environment is never ideal and thus substrate effects must be excluded to obtain relevant cell migration information. In this study, we used identically pre-treated PDMS substrates and then determined the relative activity difference of HCC cells at different growth stages. Fig. 3 is a time series of microscope images showing three different regions of a patterned PDMS surface covered with well-differentiated HCC cells (Hep-G2). No cellular migration nor any other motion of the cells was observed over the 9 hour observation period. This indicates that adhesion of the Hep-G2 cells to the surface prevented spontaneous motion. Considering that cell viability on patterned PDMS surfaces was high, as shown in Fig. 2, we attribute the inactivity observed for Hep-G2 cells to their natural character.The migration behavior of poorly-differentiated HCC cells (SK-Hep-1) on patterned PDMS substrates was also examined and results are shown in Fig. 4. We chose several adjacent cell colonies (denoted A, B, C, D, and E, in Fig. 4) to clearly show the cell migration activity. Initially, cell distribution on the PDMS surface was strongly influenced by the oxidation pattern. After 1 h, the single cell in colony A developed a pseudopod, which extended and elongated toward colony B. At 3 h, the pseudopod reached the cells in colony B forming a link between the two colonies. At 4 h, the connection site appeared wider and more robust. This movement is consistent with the first and second step (pseudopod formation and adhesion to the environment) in the multistep model of Lauffenburger and Horwitz [28]. Three adjacent multi-cells colonies (C, D, and E) were also monitored as shown in Fig. 4. No movement of these cell colonies was observed during the first five hours of the experiment. At 5 h, pseudopodia were observed in both colonies D and E, which extended toward the centroid of the three colonies. At 7 h, pseudopodia from each of the three colonies conjoined. This phenomenon was different from the pseudopod formation between cells in A and B as the connection between pseudopodia C, D, and E disconnected at 9 h and then quickly disappeared. According to the multistep model, retraction of the pseudopodia results in cell movement and a spherical cell appearance [7, 28]. However, no clear migration of cell bodies in C, D, and E was observed at 9 h, at which time the cell pseudopodia had retracted.The cell activity described above can be understood by considering the interaction from surrounding cells. For those relatively isolated cell colonies, however, activity was also observed, as shown in Fig. 5. This figure represents three isolated cell colonies, whose neighbors consist of fragmented cells. In the figure, a red line is drawn along the long axis of the cell colonies to more clearly show rotational motion. It is clear that the isolated cell colonies rotated around their centroid either counter-clockwise (Fig. 5a) or clockwise (Figs. 5b and 5c). Because no pseudopodia were formed to assist in this process, we assume the rotation was continuous, and obtained an average rotational velocity of 6.4×10 rpm byfitting the angular data from Fig. 5. Migration of the SK-Hep-1 cells on patterned PDMS was tracked over time and the results are shown in Fig. 6. Afte 24 h the sample was nearly covered by cells due to migration and redistribution. Other published also showed that observation periods of 12- 15 h were necessary to track migrating tumor cells [7, 29]. They proposed that the long observation times were necessary due to tumor cell migration rates of 0.1–0.3 μm min-1 for unstimulated cells. However, our experimental results suggest that migration of tumor cells on a patterned substrate consists of two processes,i.e. ready movement ( including pseudopodia heuristic action as shown in Fig. 4, and cell rotation as shown in Fig. 5), and cell migration (Fig. 6). Further, pseudopodia formation and movement slowed cell migration. The development of HCC cells is closely related to tumor size. Less-differentiated HCC grow within pre- existing well-differentiated HCC and may ultimately replace the well-differentiated HCC with a resulting increase in tumor size. [30]. This is a multistep process involving multiple molecular changes, and various alterations may accelerate this process. HCC cells with different histological grades represent alterations of cell properties and biological responses, including their sensitivity to medicine[31] and force [32], cellular polarity [33, 34], cell expression (adhesion molecule) [33], and transforming growth factor-α and its receptor [30]. In this study, we observed different activity between well and poorly-differentiated HCC cells, and the identification of the relation between these alterations may provide insight into the pathogenesis of HCC.Molecules presented at the apical membranes of HCC cells may play an important role in the alteration between less-differentiated and well-differentiated HCC cells. Reported results showed that < 50% of tumor cells showed markedly lower expression of the adheren junctions and desmosomes in well and moderately- differentiated HCC. In contrast, > 50% of tumor cells or nearly all cancer cells lost the ability to express adheren junctions and desmosomes in poorly-differentiated HCC [33]. Adhesion molecules in epithelial cells can maintain cellular polarity while poorly-differentiated cancer cells exhibit low polarity due to this decrease or loss of adhesion molecules. Even the polarity of hepatocytes transformed from polar to apolar during carcinogenesis [33]. Loss of cell polarity results in changes in cell distribution due to dipole arrangement, thus cells become more active.Although the two HCC cell lines (Hep-G2 and SK- Hep-1) are apolar, we only observed migration for the poorly-differentiated cells. This is closely related to the interaction between cells and the patterned PDMS substrate. Plasma patterning selectively transformed the PDMS surface from a hydrophobic (surface stoichiometry: n-Si-(CH3)2-O-n) and cell repellent state, to a hydrophilic (SiO2) and roughened state, which promoted cell adhesion and growth [24, 26]. However, molecules presented at the apical membranes of HCC cells may be one of key roles in influencing the strength of cell adhesion. For poorly-differentiated HCC cells (SK-Hep-1), a decrease or loss of these molecules, which are integral to cellular communication with substrate and neighboring cells, weakens the interaction between cells and the substrate. In contrast, well-differentiated HCC cells (Hep-G2) possess relatively more membrane molecules that can attach to the patterned PDMS substrate, thus reducing their ability to migrate, despite the driving force for redistribution due to loss of polarity.Migration of SK-Hep-1 cells was not clearly observed until 18 hours had passed (Fig. 6), indicating a non- negligible substrate adhesive force. The pseudopodia heuristic action and cell rotation was the result of a competition between spontaneous movement and substrate adhesion. In contrast to normal cells patterned on a patterned PDMS substrate, which remained stable during long-term cell culturing (>10 days), cancer cells were very active.Our study provides an in vitro experimental model for showing the activity of HCC cells. This method can further be used for in vitro static testing of cells, such as structural and mechanical property characterization, the results of which can provide valuable information for basic cell biological studies. Furthermore, our patterned polymer substrate model can be modified with different proteins or drugs for potential applications in biosensors, tissue engineering, and bioelectronics.
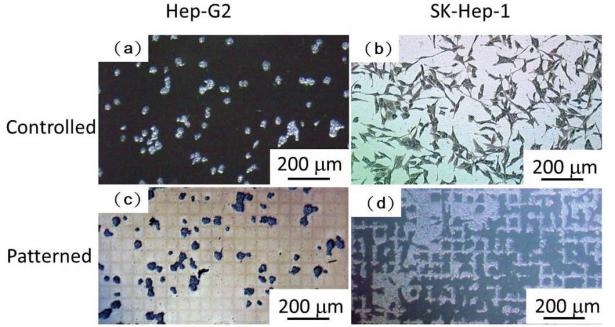
Fig. 1 Hep-G2 (a) and SK-Hep-1 (b) cells on a cover glass as a control group. Hep-G2 (c) and SK-Hep-1 (d) cells on a square patterned PDMS surface.
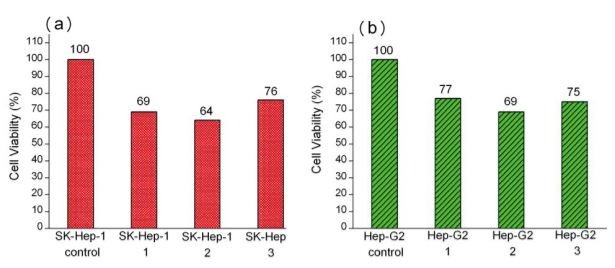
Fig. 2 Substrate dependent MTT cytotoxicity assay of (a) SK-Hep-1 and (b) Hep-G2 cells on plasma-patterned PDMS.
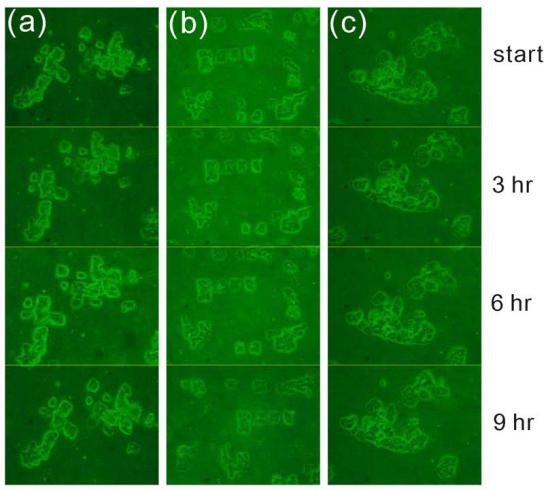
Fig. 3 Visualization of Hep-G2 cells over time. Three different locations on the surface denoted (a), (b), and (c), were monitored for 9 hours as shown.
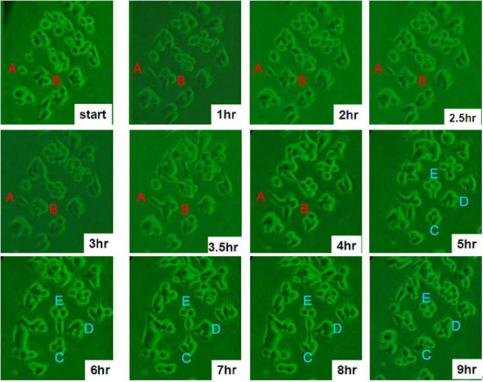
Fig. 4 Morphological changes of SK-Hep-1 cells on a plasma patterned PDMS substrate. Five characteristic cell colonies are shown denoted as A, B, C, D, and E, to clearly show cell activity.
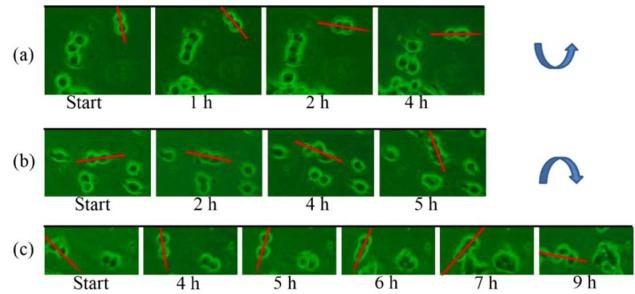
Fig. 5 SK-Hep-1 small cell colonies exhibiting (a) counter clockwise motion, and (b,c) clockwise motion. Red lines are drawn along the long axis of each cell colony to highlight the rotational movement.
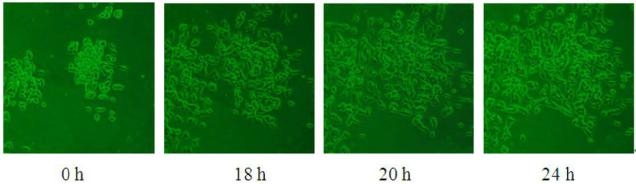
Fig. 6 Observation of SK-Hep-1 cell migration on patterned PDMS substrate after 18, 20 and 24 h durations.
In this study, we prepared patterned PDMS substrates to selectively position HCC cells of different differentiated grades, and found that the poorly-differentiated cells were more active that well-differentiated cells. Their movement consisted of ready movement (behaving as pseudopodia heuristic action and cell rotation) and cell migration. The former process was observed up to 9 h in our experiment, which is an optimal time course for some investigations requiring fixed cells. Molecules at the apical membranes of HCC cells strongly influence the cell polarity, and because they are present in relatively small quantities in less-differentiated cells, the SK-Hep-1 cells in our experiments exhibited higher mobility. In summary, we have shown that HCC cells of different differentiated grades exhibit very different migration behavior, which may be an important identifying property in pathological studies. Our model is a simple, rapid, effective, and potentially universal cell line patterning methodology for cell biology studies.
The authors thank the National Science Council (NSC 99-2113-M- 110-005 and 99-2738-M-110-001) of Taiwan and the National Sun Yat- sen University Center for Nanoscience and Nanotechnology, and the Kaohsiung Veterans General Hospital (VGHNSU100-001) for financial support of this work.
1. Matsuo N, Shiraha H, Fujikawa T, Takaoka N, Ueda N, Tanaka S, Nishina S, Nakanishi Y, Uemura M, Takaki A, Nakamura S, Kobayashi Y, Nouso K, Yagi T and Yamamoto K. Twist expression promotes migration and invasion in hepatocellular carcinoma. BMC cancer 2009; 9: 240. doi: 10.1186/1471-2407-9-240.
2. Cai AQ, Landman KA and Hughes BD. Modelling directional guidance and motility regulation in cell migration. Bull. Math. Biol. 2006; 68: 25-52. doi: 10.1007/s11538-005-9028-x.
3. Vernon RB and Gooden MD. New technologies in vitro for analysis of cell movement on or within collagen gels. Matrix Biol. 2002; 21: 661-669. doi: 10.1016/S0945-053X(02)00091-4.
4. Takagi H, Sato MJ, Yanagida T and Ueda M. Functional analysis of spontaneous cell movement under different physiological conditions. PLoS ONE 2008; 3: e2648. doi: 10.1371/journal. pone.0002648.
5. De Hauwer C, Camby I, Darro F, Decaestecker C, Gras T, Salmon I, Kiss R and Van Ham P. Dynamic characterization of glioblastoma cell motility. Biochem. Biophys. Res. Commun. 1997; 232: 267-272. doi: 10.1006/bbrc.1997.6291.
6. Yousaf MN. Model substrates for studies of cell mobility. Curr.Opin. Chem. Biol. 2009; 13: 697-704. doi: 10.1016/ j.cbpa.2009.10.001.. doi: 10.1016/j.cbpa.2009.10.001.
7. Niggemann B, Drell Iv TL, Joseph J, Weidt C, Lang K, Zaenker KS and Entschladen F. Tumor cell locomotion: Differential dynamics of spontaneous and induced migration in a 3D collagen matrix. Exp. Cell. Res. 2004; 298: 178-187. doi: 10.1016/ j.yexcr.2004.04.001.
8. Chon JH, Vizena AD, Rock BM and Chaikof EL. Characterization of single-cell migration using a computer-aided fluorescence time- lapse videomicroscopy system. Anal. Biochem. 1997; 252: 246-254. doi: 10.1006/abio.1997.2321.
9. Hou S, Yang K, Qin M, Feng XZ, Guan L, Yang Y and WangC. Patterning of cells on functionalized poly(dimethylsiloxane) surface prepared by hydrophobin and collagen modification. Bios ens . Bioelectron. 2008; 24: 912-916. doi: 10.1016/ j.bios.2008.07.045
10. Kikuchi K, Sumaru K, Edahiro JI, Ooshima Y, Sugiura S, Takagi T and Kanamori T. Stepwise assembly of micropatterned co-cultures using photoresponsive culture surfaces and its application to hepatic tissue arrays. Biotechnol. Bioeng. 2009; 103: 552-561. doi: 10.1002/bit.22253.
11. Rhee SW, Taylor AM, Cribbs DH, Cotman CW and Jeon NL. External force-assisted cell positioning inside microfluidic devices. Biomed. Microdevices 2007; 9: 15-23. doi: 10.1007/s10544-006- 9002-x.
12. Tamura T, Sakai Y and Nakazawa K. Two-dimensional microarray of HepG2 spheroids using collagen/polyethylene glycol micropatterned chip. J. Mater. Sci. - Mater. Med. 2008; 19: 2071- 2077. doi: 10.1007/s10856-007-3305-1.
13. Mori R, Sakai Y and Nakazawa K. Micropatterned organoid culture of rat hepatocytes and HepG2 cells. J. Biosci. Bioeng. 2008; 106: 237-242. doi: 10.1263/jbb.106.237.
14. Falconnet D, Csucs G, Michelle Grandin H and Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 2006; 27: 3044-3063. doi: 10.1016/ j.biomaterials.2005.12.024.
15. Borenstein JT, Terai H, King KR, Weinberg EJ, Kaazempur-Mofrad MR and Vacanti JP. Microfabrication technology for vascularized issue engineering. Biomed. Microdevices 2002; 4: 167-175. doi: 10.1023/A:1016040212127.
16. De Silva MN, Desai R and Odde DJ. Micro-patterning of animal cells on PDMS substrates in the presence of serum without use of adhesion inhibitors. Biomed. Microdevices 2004; 6: 219-222. doi: 10.1023/B:BMMD.0000042051.09807.8c.
17. Kim YC, Park SJ and Park JK. Biomechanical analysis of cancerous and normal cells based on bulge generation in a microfluidic device. Analyst 2008; 133: 1432-1439. doi: 10.1039/b805355c.
18. Zhang S, Lin Y, Altman M, Lässle M, Nugent H, Frankel F, Lauffenburger DA, Whitesides GM and Rich A. Biological surface engineering: A simple system for cell pattern formation. Biomaterials 1999; 20: 1213-1220. doi: 10.1016/S0142- 9612(99)00014-9.doi: 10.1016/S0142-9612(99)
19. Ogaki R, Alexander M and Kingshott P. Chemical patterning in biointerface science. Mater. Today 2010; 13: 22-35. doi: 10.1016/ s1369-7021(10)70057-2.
20. Wong I and Ho CM. Surface molecular property modifications for poly(dimethylsiloxane) (PDMS) based microfluidic devices. Microfluid. Nanofluid. 2009; 7: 291-306. doi: 10.1007/s10404-009-0443-4.
21. Wang L, Lei L, Ni XF, Shi J and Chen Y. Patterning bio-molecules for cell attachment at single cell levels in PDMS microfluidic chips. Microelectron. Eng. 2009; 86: 1462-1464. doi: 10.1016/ j.mee.2009.01.030.
22. McFarland CD, Thomas CH, DeFilippis C, Steele JG and Healy KE. Protein adsorption and cell attachment to patterned surfaces. J. Biomed. Mater. Res. 2000; 49: 200-210. doi: 10.1002/(SICI)1097- 4636(200002)49:2<200::AID-JBM7>3.0.CO;2-L.
23. Inoue S, Imamura M, Umezawa A and Tabata Y. Attachment, proliferation and adipogenic dif ferentiation of adipo- stromal cells on self-assembled monolayers of different chemical compositions. J. Biomater. Sci., Polym. Ed. 2008; 19: 893-914. doi: 10.1163/156856208784613541.
24. Frimat JP, Menne H, Michels A, Kittel S, Kettler R, Borgmann S, Franzke J and West J. Plasma stencilling methods for cell patterning. Anal. Bioanal.Chem. 2009; 395: 601-609. doi: 10.1007/s00216-009-2824-7.
25. Hsieh CW, Zheng B and Hsieh S. Ferritin protein imaging and detection by magnetic force microscopy. Chem. Commun. 2010; 46: 1655-1657. doi: 10.1039/b912338e.
26. Hsieh S, Cheng YA, Hsieh CW and Liu Y. Plasma induced patterning of polydimethylsiloxane surfaces. Mater. Sci. Eng., B 2009; 156: 18-23. doi: 10.1016/j.mseb.2008.10.036.
27. Ertel SI, Ratner BD, Kaul A, Schway MB and Horbett TA. In vitro study of the intrinsic toxicity of synthetic surfaces to cells. J. Bi ome d. Mate r. Re s. 1994; 28: 667-675. doi : 10.1002/ jbm.820280603.
28. Lauffenburger DA and Horwitz AF. Cell migration: A physically integrated molecular process. Cell 1996; 84: 359-369. doi: 10.1016/ S0092-8674(00)81280-5.
29. Niggemann B, Maaser K, Lü H, Kroczek R, Zänker KS and Friedl P. Locomotory phenotypes of human tumor cell lines and T lymphocytes in a three-dimensional collagen lattice. Cancer Letters 1997; 118: 173-180. doi: 10.1016/S0304-3835(97)00328-5.
30. Morimitsu Y, Chu Chieh H, Kojiro M and Tabor E. Nodules of less- differentiated tumor within or adjacent to hepatocellular carcinoma: Relative expression of transforming growth factor-α and its receptor in the different areas of tumor. Human Pathology 1995; 26: 1126- 1132. doi: 10.1016/0046-8177(95)90275-9
31. Chiang PC, Lin SC, Pan SL, Kuo CH, Tsai IL, Kuo MT, Wen WC, Chen P and Guh JH. Antroquinonol displays anticancer potential against human hepatocellular carcinoma cells: A crucial role of AMPK and mTOR pathways. Biochem. Pharmacol. 2010; 7 9: 162-171. doi: 10.1016/j.bcp.2009.08.022.
32. Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F and Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stemcells. Nat. Mater. 2010; 9: 82- 88. doi: 10.1038/nmat2563.
33. Cao Y, Chang H, Li L, Cheng RC and Fan XN. Alteration of adhesion molecule expression and cellular polarity in hepatocellular carcinoma. Histopathology 2007; 51: 528-538. doi: 10.1111/j.1365- 2559.2007.02820.x.
34. Cao Y, Schlag PM and Karsten U. Immunodetection of epithelial mucin (MUC1, MUC3) and mucin-associated glycotopes (TF, Tn, and sialosyl-Tn) in benign and malignant lesions of colonic epithelium: apolar localization corresponds to malignant transformation. Virchows Archiv 1997; 431: 159-166. doi: 10.1007/ s004280050083.
Copyright:(c) 2011 B. Zheng, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
![]()