Review
Microfluidic PCR Chips
Jingdong Chen 1, Di Chen 1*, Tao Yuan 1, Xiang Chen 2
1 National Key Laboratory of Science and Technology on Micro/Nano Fabrication Technology.
2 Key Laboratory for Thin Film and Microfabrication of the Ministry of Education
Research Institute of Mico/Nano Science and Technology, Shanghai Jiao Tong University, Shanghai 200240, China.
* Corresponding author. Email: dchen@sjtu.edu.cn
Citation: J. Chen et al. Microfluidic PCR chips. Nano Biomed. Eng. 2011, 3(4), 203-210.
DOI: 10.5101/nbe.v3i4.p203-210.
The microfluidic polymerase chain reaction (PCR) chips have undergone extensive development and nowadays have become an important domain of miniaturization technology application. Here, we review the advances of microfluidic PCR chips over the past years, from the first single chamber stationary PCR chip to the new SlipChip PCR. First, the three distinct types of microfluidic PCR chips are discussed, including chamber stationary PCR chips, flow-through PCR chips and convection PCR chips. Then we focus on droplet PCR chips and SlipChip PCR. Although they are at an early stage, they show the great potential for high-throughput PCR and robust chip. Finally, general discussions on integrated chips are given. The low cost, portable, high-throughout integrated PCR chips will certainly be further developed in spite of many challenges.
Keywords: Chamber stationary PCR chip, Flow-through PCR chips, Convection PCR chip, Droplet PCR chip, SlipChip PCR, Integrated PCR chip
1. Introduction
Since micro total analysis system (μ-TAS), also known as “lab-on-a-chip” was proposed in the early 1990s [1], microfluidic chip manufactured by microelectromechanical system (MEMS) technology has been considered as a potential technology to miniaturize the conventional equipments and technologies. Because it offers advantages in terms of small volume, low cost, short reaction time, high throughput. It has already been used in chemical and biological analysis [2,3], cell analysis and clinical diagnostics [4-6] drug discovery [7], and environmental monitoring [8]. Among the microfluidic chips, the polymerase chain reaction (PCR) chip has become a very important tool in modern biology, biomedical research and related areas. The PCR technique was first developed in 1985 [9], and has been widely used as a molecular biological tool to replicate DNA by cycling through three temperature steps. After the first PCR chip was introduced by Northrup et al. [10], many research groups began to study microfluidic PCR chips and developments of microfluidic PCR chips were accelerated. This review surveys the development of different microfluidic PCR chips. We begin with the three distinct types of microfluidic PCR chips (chamber stationary PCR chips, flow-through PCR chips and thermal convection- driven PCR chips). Next, we describe droplet PCR chips and SlipChip PCR with the great potential for high-throughput PCR and robust chip. Finally, the potential and challenges about integrated PCR chips are discussed. As a supplement to this review, readers are referred to other reviews about microfluidic technology [11-14].
2. Types of Microfluidic PCR Chips
During the development of microfluidic PCR chips, different types have been developed by many research groups. Currently, the microfluidic PCR chips can be classified into three distinct types: chamber stationary PCR chips, flow-through PCR chips and thermal convection-driven PCR chips.
The working principle of this type of PCR chip is that the PCR solution is kept stationary and the temperature of the reaction chamber is cycled between three different temperatures. In general, this type of chamber stationary PCRchips can be classified as single chamber [15-17] and multi-chamber [18-20] chips, as shown in Fig.1. The first PCR chip developed by Northrup et al. [10] was based on a chamber fabricated by silicon anisotropic wet etching, where the PCR solution is kept. Cady et al. [21] constructed the PCR amplification chamber using soft lithography techniques for polydimethylsiloxane (PDMS) and SU-8 photoresist tested for their ability to purify and detect the pathogeneic bacterium Listeria monocytogenes Qiu et al. [22] developed a disposable, plastic microfluidic reactor with relatively large reaction volume (ranging from 10 μL to 100 μL) and used double- sided heater to maintain temperature uniformity and a relatively fast temperature ramping rate. The fluidic and thermal controls can be performed very well. However, a single chamber is not suitable for some sequential PCR tests for a quantity of DNA samples. In order to solve this problem, multi-chamber stationary PCR chips was developed. Daniel et al. [23] fabricated some microchambers in silicon by bulk micromachining using anisotropic wet etching and integrated thin film platinum resistors as temperature sensors and heaters to amplify DNA. Multi-chamber PCR chip for multi-target sample amplification for diagnostic purposes was designed and fabricated by Trung et al. [24]. Silicon-based microchamber array was used to realize high-throughput PCR and PCR was completed in 18 min for 40 cycles [25]. The multi-chamber stationary PCR chips can reduce the time for analysis and increase the PCR throughput because it can perform different sequential PCR tests concurrently. However, it is crucial to ensure temperature uniformity between chambers. The whole chip, including the sample, is heated and cooled through specific thermal-cycling temperatures. Therefore, chamber stationary PCR chips have high thermal inertia and long thermal-cycling time.

Fig. 1 Chamber stationary PCR chips: (a) Single chamber PCR chip.(.b) Multi-chamber PCR chip [12].
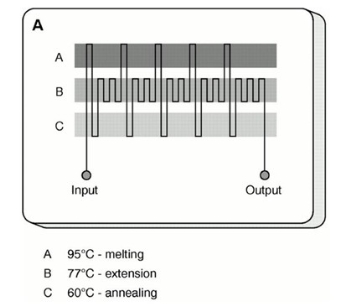
Fig. 2 Flow-through PCR chip [26].
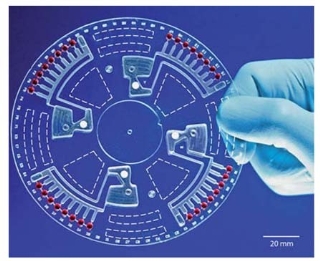
Fig. 3 Photograph of the centrifugal microfluidic chip designed by Focke et al. [33].
The working principle of this type of PCR chip is that the PCR solution is continuously and repeatedly flowing through three different temperature zones necessary for DNA amplification, as shown in Fig.2. It takes advantage in term of rapid heat transfer and high potential for further integration. Kopp et al. successfully performed the polymerase chain reaction (PCR) in continuous flow at high speed using a micromachined chemical amplifier was successfully [26]. The device relied on the movement of sample through thermostated temperature zones on a glass microchip. Since then, flow-through PCR chips have undergone substantial improvements. In this chip, the flow control of the PCR solution is a key issue.
Pressure driven is commonly used by many research groups, which control fluid movement with different pressures applied by mechanical pumps or gas pressure. Liu et al. [27] developed a rotary microfluidic device to run the PCR in both spatially and temporally cycled formats. The small sample volume (12 nl) allows low power consumption, reduced reagent costs, and ultimately more rapid thermal cycling. Wu and Lee [28] proposed a three-dimensional (3D) on-chip continuous-flow PCR, which adopted a single heating source to simplify the temperature control.
Electrokinetic force driven is another important fluid control technique in microfluidic chips [29]. Patankar and Hu [30] developed a numerical scheme to simulate electroosmotic flows in complicated geometries and studied the electroosmotic injection characteristics of a cross-channel device for capillary electrophoresis. Gui and Ren [31] proposed a 3D model to simulate the electrical potential field, the flow field, and the temperature field in an electroosmosis-based continuous flow PCR chip.
Centrifugally force driven is an insightful example of the ingenious methods for fluid control which may be used in microfluidic chips. Instead of mechanical or electrokinetic pumps, centrifugal force is used which moves liquids inside circular disks containing all necessary microfluidic components for chemical analysis. Furutani et al. [32] proposed a compact disk (CD)-shaped chip to isolate Salmonella enterica cells and detect the Salmonella- specific invA gene from isolated cells by PCR.The centrifugal force was used to control liquid flow, without a micro-pump. Focke et al. [33] developed a centrifugal microfluidic chip and successfully and realized efficient thermocycling during real-time PCR, asshown in Fig. 3.

Fig. 4 (a) Schematic drawing of the PCR microchip with heating blocks and a magnet, (b) Photograph of the PCR chip fabricated in PMMA [35].
In this chip, a small ferrofluid plug is controlled by an external magnet, which in turn propels the PCR solution through three temperature zones. Sun et al. [34] presented a novel circular close-loop ferrofluid microfluidic chip for rapid PCR and successfully demonstrated by performing PCR amplification of a 500 bp lambda DNA fragment and a 16-loci forensic DNA sample. Then they amplified the genetically modified soya and maize in less than 13 min for the detection of genetically modified organisms (GMOs) in food products using PCR technology, as shown in Fig. 4 [35]. The high thermal inertia has been dramatically shortened by flow-through PCR chips. However, PCR inhibition and contamination become the major challenges due to high surface/volume ratio [36, 37].
The working principle of convection PCR chip is Rayleigh-Bénard convection, which is caused by buoyancy-driven instability in a confined fluid layer heated. This convection PCR chips consist of two fixed different temperature zones. Buoyancy force is the only force to drive PCR solution flow through the temperature zones. Krishnan et al. [38] reported a PCR convection system to perform PCR amplification of DNA inside a 35-μL cylindrical cavity. The temperature cycling was generated as the flow continuously shuttles fluid vertically through the two temperature zones of denaturation (97°C) and annealing/extension (61°C). Yao et al. [39] designed a micro-PCR system that integrated measurement circuits, and circuits to control the temperature. They optimized the stream function, velocity, and temperature profile through simulation and found the minimum reaction duration as a function of the velocity, temperature distribution, and extension time for a specific aspect ratio of the convection cavity. Chung et al. [40] presented a standalone portable convection PCR chip which can conduct multiple PCR in a very short time (about 5 min). Natural convection drove PCR reactants to circulate along a closed loop channel in the polymer chip, as shown in Fig. 5. Convection PCR is predicted to be a promising method due to its low cost, simplicity, high speed and low power consumption [41, 42]. And challenges still exist in realization of parallel and multiplex convective PCR and integration of convective PCR with other functionalities.
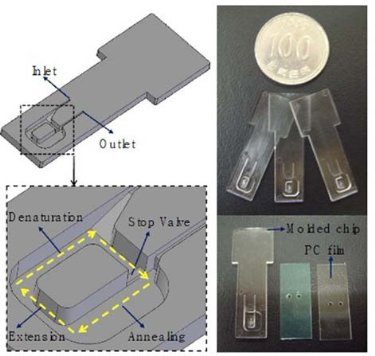
Fig. 5 .Convection PCR chip (a) design, (b) fabrication [40].
3. Droplet PCR Chip
In order to prevent PCR inhibition and contamination, some researchers use surface modification method [43- 45]. Schneegass et al. [43] modified the channel surface with hexamethyldisilazane (HMDS) before the injection of the PCR solution, so the hydrophobic material surface of the silicon/glass was changed to enhance surface biocompatibility. However, this method is not easy to manipulate in the sealed chip. Fortunately, the PCR inhibition and contamination can be overcome by using droplet PCR chip. In general, droplets are typically generated using three main generation strategies: capillary, T-junction and flow focusing, as shown in Fig. 6 [46]. In droplet PCR chip, the PCR solution (water phase) within the droplets are surrounded by an immiscible fluid (oil phase), so the inhibition and contamination result from contact between chambers and the sample is avoid. In addition, The PCR solution contained in droplets can decrease its evaporation, which is a common problem in single-phase chips. Unlike in single-phase chips, each independent droplet in droplet PCR chips likes a microreactor which can be individually transported, split, mixed, sorted and analyzed [46-53]. Pipper et al. [54] used magnetic force to manipulate a free droplet containing superparamagnetic particles to detect the highly pathogenic avian influenza virus H5N1 in a throat swab sample. The RT-PCR results showed equally sensitive, 440% faster and 2,000-5,000% cheaper compared to commercially available tests. Wang et al. [55] designed and fabricated a droplet-based micro oscillating flow PCR chip by the silicon microfabrication technique to miniaturize the flow process while maintaining the advantages of fixed temperature conditions. The results demonstrated that the chip successfully amplified the HPV-DNA, with a processing time of about 15 min. Mohr et al. [56] investigated some droplet PCR chip design factors including thermal mass, flow rate and thermal resistance. Specially, they focused on the fluid and substrate temperature distribution within the PCR chip and the droplet residence times in critical temperature zones. Some parameters related to PCR amplification efficiency were studied, such as reagent concentration, droplet size and hold time at each temperature step [57]. The droplets, as individual reactors, can contain many DNA templates and be amplified individually. In addition, droplets can be produced at a very high frequency within one experiment, so parallel processing is achievable to produce large data sets and offer higher degree of confidence following analyses [58]. Droplet chips serve as an ideal mean to a future generation of high-throughput droplet PCR [60-64]. A high-throughput microfluidic chip that encapsulates PCR reagents in millions of picoliter droplets in a continuous oil flow was developed and it showed high sensitivity (detection of template concentrations as low as 0.003 pg/μL in 35 min)[65]. Srisa-Art et al. [66] demonstrated a novel, high-throughput, droplet-based microfluidic assay and allowed for online characterization and detection of droplets (with typical volumes of 300 pl) at rates in excess of 1 kHz. Andrew et al. [67] proposed a high-throughput microfluidic chip, which is capable of generating over 1-million, monodisperse, 50 pl droplets in 2-7 minutes, as shown in Fig. 7. In addition, the chip was integrated rapid droplet generation, thermocycling, and wide-field fluorescence imaging. Although the development of droplet PCR chips is still in their infancy, they have shown the great potential, particularly for high-throughput PCR [60-64].
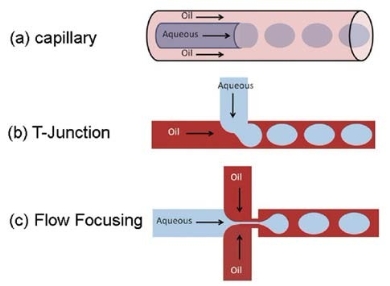
Fig. 6 .Droplet generation strategies: (a) co-flow in a capillary format, (b) T-Junction in a planar chip format, (c) flow focusing in a planar chip format [46].
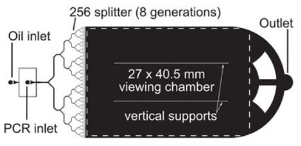
Fig. 7 .High-throughput microfluidic PCR chip [67].
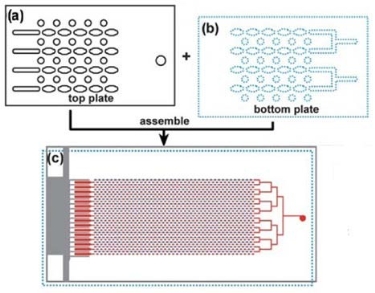
Fig. 8 Schematic drawing of digital SlipChip PCR for: (a) part of the top plate, (b) part of the bottom plate, (c) the entire assembled SlipChip after slipping. [74]
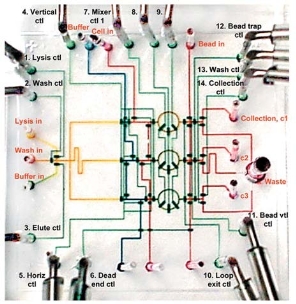
Fig. 9. Integrated PCR chip [80].
Recently, the team in the Uiversity of Chicago pioneered an ingenious method to perform microfluidic chip without external pumps, valves and other equipment for operation. The system consists of two glass plates with arrays of wells and channels. By moving one plate relative to another plate, reagent and sample can be brought into contact and reaction [68-70]. The SlipChip has been applied to protein crystallization [71, 72], immunoassay [73], nucleic acids analysis [74-76]. They proposed a very simple and inexpensive digital SlipChip PCR that contained 1280 droplets of 2.6 nl each, and was capable of detecting the template DNA at single copy level [74]. Then they developed a high- throughput nanoliter multiplex SlipChip PCR with robust performance and lack of false negatives, false positives, and cross-contamination. The chip was designed to preload one primer pair per reaction compartment and to screen up to 384 different primer pairs with less than 30 nl of sample per reaction compartment [75]. The SlipChip PCR can perform multiplex detection with different primer pairs loaded in different reaction chambers. Although SlipChip is at an early stage of its development, it offers a new microfluidic platform that opens up new possibilities for the realization of robust chip laboratories [70].
The PCR chip has undergone the transition from simple microfluidic components to highly integrated systems. An ideal integrated PCR chip can integrate individual microfluidic components, such as cell isolation and capture, cell lysis, DNA/RNA extraction and purification, DNA/RNA amplification, and product detection into a single automated, portable chip with sample-to-answer capability. It is attractive and challenging to realize the integrated chip, especially the low cost, portable, high- throughput integrated chip. Yuen et al. [77] isolated white blood cells from whole blood by filter section of the microchip and subsequently directly performed PCR. The microchip provides a convenient means to simplify nucleic acid analyses by integrating two key steps: cell isolation and PCR. Ferrance et al. [78] reported a microchip integrated extraction of genomic DNA, IR-mediated PCR amplification and electrophoretic analysis. They used a novel sol–gel matrix to extract of genomic DNA from whole blood, and detected directly by electrophoretic analysis in the chip. Liu et al. [79] developed a self-contained, fully integrated biochip. The chip started with magnetic bead-based target cell capture, cell concentration and purification, and cell lysis, followed by PCR amplification and electrochemical DNA microarray-based detection. The chip provides a cost-effective solution to direct sample-to-answer genetic analysis. Hong et al. [80] developed microfluidic chips for automated nucleic acid purification from small numbers of bacterial or mammalian cells. The chip has 26 access holes, 1 waste hole and 54 valves within 20 × 20 mm2, as shown in Fig. 9. The chip involves all processes in term of cell isolation, cell lysis, DNA or mRNA purification, and recovery, so any pre- or postsample treatment is not needed. In addition, the chips are capable of processing different samples in parallel, thereby illustrating high- throughput performance. The PCR product analysis is also important in the integration chip. Some reviews have presented this issue in detail [81, 82]. One of the main challenges in miniaturization is the integration of functional components to perform several operations without the need for macro apparatus or manual user input. Higher levels of integration can reduce or avoid cross-contamination in a multitude of samples to be run in parallel [83-86].
The microfluidic PCR chips have undergone extensive development and dramatically changed conventional PCR for DNA amplification. Since the first PCR chip was developed, many microfluidic PCR chips with various characteristics were designed to meet challenges. Chamber stationary PCR chips have high thermal inertia and long thermal-cycling time. Flow-through PCR chips have short cycling time, but result in PCR inhibition and contamination. Convection PCR is predicted to be a promising technology, but challenges still exist in realization of parallel and multiplex convective PCR and integration of convective PCR with other functionalities. To overcome the drawbacks in Flow-through PCR chip, droplet PCR chips are used widely. In addition, they have shown the great potential, particularly for high- throughput PCR, although they are still in the early stage. The ingenious SlipChip PCR provides a method for multiplex detection and opens up new possibilities for the realization of robust chip laboratories. Unfortunately, few microfluidic PCR chips have been commercialized. But there is no doubt that low cost, portable, high-throughout integrated PCR chips will be widely used in the future.
This work was supported by Ministry of Science and Technology of China (2010CB933901), Science and Technology Innovation Fund of SJTU - University of Michigan.
Copyright:(c) 2011 J. Chen, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.