![]()
Article
2D Gold Nanoparticle Structures Engineered Through DNA Tiles for Delivery and Therapy
Ali Yasin Sonay, Kemal Keseroğlu and Mustafa Culha*
Department of Genetics and Bioengineering, Faculty of Engineering, Yeditepe University, Ataşehir, Istanbul 34755, Turkey.
*Corresponding author. Email: mculha@yeditepe.edu.tr
Citation: Ali Yasin Sonay, et al. 2D Gold Nanoparticle Structures Engineered Through DNA Tiles for Delivery and Therapy. Nano Biomed. Eng. 2012, 4(1), 17-22.
DOI: 10.5101/nbe.v4i1.p17-22.
Abstract
Self-assembly of 13 nm gold nanoparticles (AuNPs) engineered into 2D structures in solution using DNA tiles for their possible use for gene delivery and photothermal therapy is reported. The two different DNA tiles were constructed and the AuNPs coated with oligonucleotides possessing complementary sequence from the free ends were hybridized with the sticky ends of the tiles. The DNA tiles were bind to each other by mixing the tile structures without a heating and cooling step. The constructed nanostructures were 5 to 7 DNA tiles long since heat was not used to elongate them. When the DNA tiles were bound to the AuNPs, it was observed that AuNPs tend to stay in close proximity by filling the gaps between tiles. The stability of the constructed structures was tested against DNase, a DNA cleaving enzyme, for possible applications for gene delivery and photothermal therapy. It was found that the AuNP bound DNA tile structures resist the DNase cleavage up to eighty percent. Due to the presence of the AuNPs in the structure, the enzyme cannot bind to DNA sequences and this increases the DNA tile structures’ stability..
Keywords: Self-assembly, DNA Tile, Gold nanoparticles, Delivery and Photothermal therapy;
1. Introduction
Assembly of nanoparticles into 2D or 3D organizations is one of the prevalent dreams in nanoscience and nanotechnology. However, the assembly of nanostructures continues to be one of the biggest challenges in nanoscience and nanotechnology and achieving such a goal has critical importantance to take full advantage of extraordinary physicochemical properties of those novel nanomaterials. The top down approach is routinely used for bringing the nanoparticles into the patterns with some degree of successes using various lithographic techniques.1,2 However, most of top-down approaches are time consuming and require expensive instrumentation, most of the time, along with trained personnel.3 Therefore, search for novel routes employing bottom up approaches has gained importance in recent years.4,5 The main venue in bottom up approaches is programmed self assembly, which relies on the well-defined interactions between natural macromolecules like DNA,6 RNA,7 peptides,8 which are used to construct nanostructures. Among those macromolecules, the most promising one is DNA because of its meta-stable structure, biocompatibility and well-defined hybridization property. Several techniques have been applied to use DNA in the assembly of nanostructures.9-17 One such technique is DNA origami, using Watson-Crick base pairing, constructing DNA structures with desirable shapes. In this strategy, large DNA structures are engineered by folding of a single stranded long DNA with a single stranded smaller DNA (staple strands). By using DNA Origami, it is possible to construct 2D DNA shapes; such as star, smiley faces and triangles.18 In 2003, the group of LaBean and Yan designed 4x4 DNA tiles and constructed 2D grids by assembling the tiles.19 Seaming of DNA Origami is also possible, which helps to produce 3D shapes from DNA. In the past years, Andersen et al. constructed a 3D DNA box with a controllable lid20 and Sharma et al. perapared DNA tubules.21 The AuNPs offer unique opportunities for development of novel therapeutic and delivery agents for diagnosis and disease treatment due to their easy surface chemistry and tunable optical properties. The AuNPs can both absorb light and scatter simultaneously22. While scattered light is used for imaging, the absorbed light, which is converted to heat, can be used for photothermal therapy23. The scattering or absorbing properties can be tuned by changing the aspect ratio, which is defined as the ratio of the length to the diameter, and inter particle distance in the case of aggregates23-25. For example, the AuNP size and shape can be tuned to enhance the light adsorption in the NIR region of the spectrum to decrease the damage of the radiation to the health cells or tissue. Several reports demonstrated the proof of concept of heat-based killing of cancer cells using AuNPs26, 27. When the AuNPs are modified either with an antibody or aptamer for selective targeting, they selectively bind the target cell surface. The construction of multipurpose higher order structures using the AuNPs is possible. For example, a 2D construct using the AuNPs as building blocks for not only thermaltherapy but also delivering a cargo such as a drug or gene can be designed. This structure is transfected into the cells selectively to deliver their cargo and to be used for heat killing as better-defined structures other than just regular aggregates. The studies for assembly of nanoparticles with DNA origami were also reported.28-34 The DNA tiles have been used to construct several nanogrids30 and small sized AuNPs bound to oligonucleotides that were hybridized with this nanogrid on the surface. In our study, we used DNA tiles with 13 nm AuNPs to assemble 5-7 tiles long AuNP constructs. Instead of creating a nanogrid, we hybridized DNA tiles with AuNPs at the upper and lower arms. The left and right arms of tile have been used to elongate the tile structure. In the proposed model, two different DNA tiles can bind to each other and form a linear shaped structure that 13 nm AuNPs can bind from the sides. While previous attempts created nanostructures on surfaces for electrical and optical applications, our approach provides a structure that can be used for not only heat killing but also delivery of drugs or genes since the oligonucleotides coating the AuNP surface can be used for further attachment of a cargo.
2. Experimental Section
The DNA sequences used to construct the structures are provided in the supporting information (Table 1 and 2). The DNA tiles were prepared in the TAE/Mg2+ buffer by using slow cooling method. Oligonucleotides were mixed in 200 μL solution so that their final concentration would be 1.0 μM. A 2 liter of water was heated to 95 oC and placed into a styrofoam box. Oligonucleotide mixture was placed into the box and cooled down for approximately 40 hours. Then, to strengthen the hybridization, the samples were placed into +4 oC overnight.30 The 13-nm size of spherical AuNPs were synthesized using citrate reduction method35 First, a 0.2 g of HAuCl4.3H2O was dissolved in 500 ml distilled water in a flask cleaned with dichromic acid solution. This solution was heated and stirred until boiling. After boiling, a 50 ml of 38.8 µM sodium citrate solution was added slowly. This solution was kept boiling for 15 minutes. After cooling, the colloidal suspensions were filtered with 0.45 µm pore size filter, Millex (33 mm) Filter Unit with MF-Milliapore Membrane. The average size distribution of the sample was measured in Zetasizer as 13 nm and observed under TEM. Figure 1A and B shows the UV/Vis Spectroscopy spectrum and TEM images of the synthesized AuNPs. Dynamic Light Scattering (DLS) measurements were performed using a Zetasizer nano-ZS from Malvern Instruments (Malvern, UK). Each sample was measured three times and average size was determined by number for synthesized AuNPs which was 13 nm as can be seen from the Figure 1C. The TEM measurements were performed with a JEOL-2100 HRTEM operating at 120 kV and 200 kV (LaB6 filament) and equipped with an Oxford Instruments 6498 EDS system. Copper TEM grids coated with carbon support film were used to analyze samples upon placing very small drops of samples onto them. The UV/Vis spectra of the structures were recorded using a Perkin Elmer Lambda 25 UV-Vis Spectrophotometer. The concentration of DNA before and after DNase treatment was measured by Implen NanoPhotometer™ Pearl in its dsDNA mode.
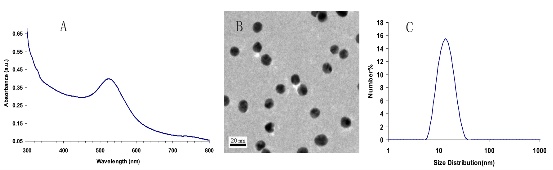
Figure 1. UV/Vis Spectrum (A), TEM image (B) and DLS spectrum (C) showing size distribution in suspension of 13 nm AuNPs.
3. Results and Discussions
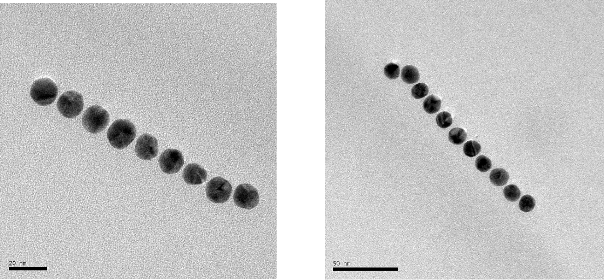
The illustrations of the structures aimed to construct is provided in Figure 2. First, the DNA tiles were redesigned based on the structure in the study of LaBean and Yan.19 The edge of upper and lower arms of the tiles were constructed with ten single stranded Adenine bases, and hybridized with Thymine bases that are bound to 13 nm AuNPs via thiol groups at the edge of the oligonucleotides. The two different DNA tiles have been created, named Tile A and Tile B. Tile A can bind to Tile B from its sticky ends, and thus forming elongated multiple tiled structures. Previously, in the literature, Tile A and B were bind to each other by slow cooling from 42 oC to 38 oC, but in our study, they were just mixed and placed on a shaker overnight. It was observed through TEM images that, tile structures tend to limit their size and form tile complexes consisting of 5-7 DNA tiles. Next, the AuNPs coated with oligonucleotides that are complementary to the upper arms of the DNA tiles were hybridized with single DNA tiles. Before hybridizing Tile A and B with each other, the AuNPs were hybridized with Tile A and Tile B in separate test tubes. After completing this hybridization with both Tile A and Tile B, they were hybridized with each other. In one case, tiles were designed in a way that only upper arms of DNA tiles contain free Poly A group that can bind to Poly T modified AuNPs. In another case, tiles were designed in a way that both upper and lower arms of DNA tiles contain free Poly A group and AuNPs can bind on both sides. Figure 3 shows UV/Vis spectra of these structures in suspensions compared to the AuNP containing suspension. The intensity of the typical band seen with the suspensions containing 13 nm AgNPs at 260 nm increases with constructed structures. The increase in the intensity of this band is attributed to the double stranded DNA. The band at 520 nm is typical for AuNPs, but as they form the assembly, this band shifts to higher wavelengths, 530 and 540 nm, for the AuNPs combined with tiles on their both arms, respectively. A small band at around 760 nm is also observed due to the possible larger structures formed. Figure 4 shows the size distribution of AuNPs compared to tiles that contain AuNPs on their upper arm and on both arms. The results indicate that AuNP size is around 13 nm whereas the other structures has sizes around 80 nm and 180 nm respectively, which is in agreement with the proposed model where tiles are lined up at sides of 5-7 DNA tiles. Although larger structures are observed with DLS, only a small wavelength shift (about 10 nm) is observed on the UV/Vis spectrum of the suspensions containing the assembled structured. This possibly due to the greater distance between the AuNPs, which the surface plasmons cannot effectively couple for absorption at higher wavelengths. The gap size should be smaller than 4 nm between the noble metal nanoparticles for effective surface plasmon coupling.37 To observe the orientation of these tiles, TEM images were taken. Figure 5 shows the TEM images of the constructed structure. As can be realized from 4 nm gaps between AuNPs, there is a DNA strand between keeping them apart. Also in this structure, because AuNPs are coated with highly negatively charged oligonucleotides, AuNPs are forced inside the gaps between DNA tiles. Figure 5A shows 9 AuNPs indicating there is 5 DNA tiles lined up to create a linear array. In this array, the gap between AuNPs is around 4 nm. The same conclusion can be drawn from Figure 5B, which has 11 AuNPs and 6 DNA tiles, again with about 4 nm gap between AuNPs. Finally, AuNPs were hybridized with DNA tiles from both sides. Theoretically, four AuNPs must be attached to each tile from both sides and the AuNPs are expected to form two linear arrays next to each other with a 4 nm gap. Figure 6 shows the TEM images of the prepared structures. On Figure 6A, the AuNPs form the expected structure. However, Figure 6B shows that irregular structures composed of the DNA tiles are also possible. Figure 6C shows eight AuNPs, four on top of the other four. The structure has a missing AuNPs on upper array. This indicates the hybridization of AuNPs to DNA tiles is not completely efficient. The use of oligonucleotide coated AuNPs for gene delivery was demonstrated previously.39 Since such 2D engineered structures can be used for gene or drug delivery with further modifications through the free oligonucleoties residing on AuNPs, the stability of these Tile-AuNP structures were tested for their stability against DNase attack, which is one of the major problems associated with the use of such structures for delivery.38 The engineered 2D structures were treated with 1 unit/μl DNase. The initial DNA concentration in the solution where tile structure formed without AuNPs was measured as 160 ng/mL. The initial DNA concentration after the tile structures treated with the AuNPs was measured as 130 ng/mL. The mixtures of DNA tile and DNase were incubated at 37 oC for 5 minutes and 0.05 M EDTA solution was added to stop the reaction. Tile without AuNPs was used as a positive control. Also two structures that are previously shown, DNA tile containing AuNPs on the upper arm and DNA tile containing AuNPs on both arms were tested. Since AuNPs give absorbance at 260 nm AuNP solution was used as blank to prevent false positives. The results of DNase treatment are given in Figure 7, where DNA concentration is shown before and after DNase treatment for these three structures. As can be seen from Figure 5B, the Tile without AuNPs was degraded 80 percent. On the other hand, the Tile containing AuNPs only on the upper arm is degraded 60 percent since AuNPs protect the upper arm, but not the lower arm. Yet when AuNPs are bound to both arms only 23 percent of the DNA is damaged which indicates that the AuNPs’ position on DNA tile protects it from DNase cleavage. Also the mixture of DNA tiles and AuNPs without hybridization was tested to see if the resistance against DNase treatment is a result of AuNP inhibition rather than the specific shape of constructed structures. It was shown that this mixing of DNA tiles and AuNPs doesn’t prevent the degradation of DNA tile structures.
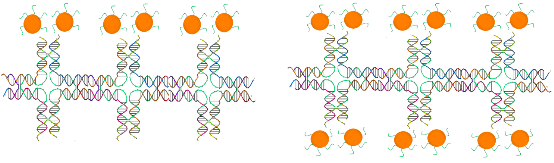
Figure 2. Two AuNPs on top of DNA tiles (A) and four AuNPs on a single DNA tiles (B), two on top, and two at the bottom. These figures are not in scale to show the hybridization of AuNPs and DNA tiles
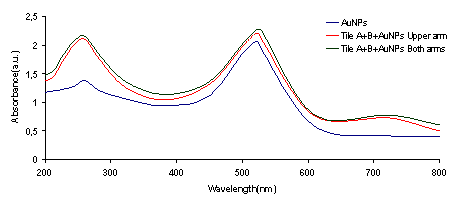
Figure 3. UV/Vis spectra of AuNPs compared to Tile-AuNP structures
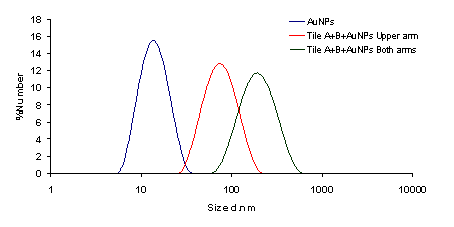
Figure 4. Size distribution of AuNPs compared to tile-AuNP structures
![]()
![]() Figure 5. 9 AuNPs lined up on 5 DNA tiles (A) and 11 AuNPs lined up on 6 DNA tiles (B).
Figure 5. 9 AuNPs lined up on 5 DNA tiles (A) and 11 AuNPs lined up on 6 DNA tiles (B).
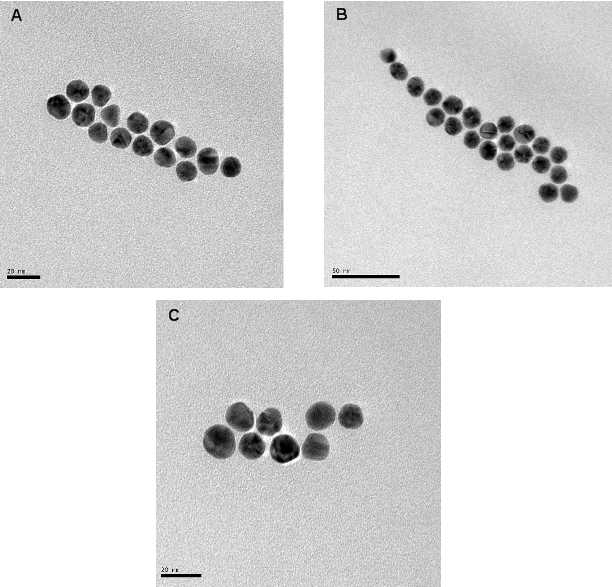
Figure 6. AuNPs forming a linear structure, consisting of two arrays (A). Triple array with different array lengths, yet protecting the interparticle gap between them (B). Formation of two linear arrays with 8 AuNPs (C).
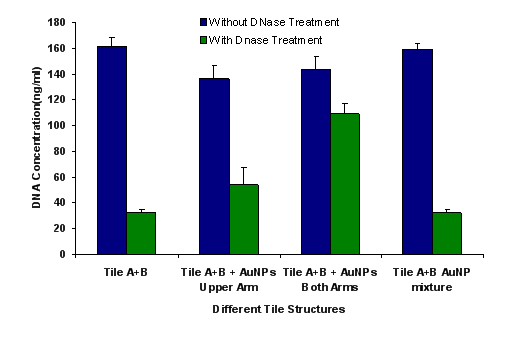
Figure 7. DNase treatment of prepared tile structures
4. Conclusions
Our results show that in contrast to approaches that are available in the literature (slow cooling from 42 oC to 38 oC and +4 oC overnight) when DNA tiles are incubated without heating and when AuNPs are hybridized with tiles before Tile A and B are hybridized with each other, they form linear tile structures ranging between 5 to 7 DNA tiles long. We hybridized two AuNPs on top of DNA tiles and the other two at the bottom. It is shown that AuNPs pack into the gaps between two hybridized DNA tiles, forming single, double and triple arrays with about 4 nm gaps between two AuNPs. The major limitation with the use of DNA based structures in biological applications is their lack of stability under enzymatic attack.38 This study demonstrates that the AuNPs protect the DNA from enzymatic cleavage since they occupy the binding sites of DNases. In conclusion, we show an alternative approach that the smaller tile structures can be created and applied for thermal killing, delivery and targeting. Since AuNPs provide additional binding sites, a variety of DNA or RNA sequences can be hybridized with them and carried along with DNA Tile-AuNP structures. For example, aptamers prepared for targeting can be used to direct the tile structures to the desired target. Although we only demonstrated the construction of the 2D structures and their enzyme resistance, we continue to explore the possible use of these structures for photothermal therapy, gene delivery and drug delivery.
Acknowledgements
The authors greatly acknowledge the financial support of TUBITAK (Grant no:108T605) and Yeditepe University. We also thank Dr. Özgür Duygulu for his help on TEM images.
References
Copyright: (c) 2012 AY, Sonay, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.