Research Article
Dendrimer-Modified Gold Nanorods as High Efficient Controlled Gene Delivery Release System under Near-Infrared Light Irradiation
Chenlu Li 1, Fangfang Xia 2, Kan Wang 2, Can Wang 2, Ping Xu 2, Hong Zhang 2, Jinping Wang 2, Asahi Toru 2, Jian Ni 2*, Daxiang Cui 1, 2, 3*
1 Key Laboratory of Laboratory Medicine, Ministry of Education of China,Zhejiang Provincial Key Laboratory of Medical Genetics, School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, Zhejiang 325035, P. R .China.
2 Institute of Nano Biomedicine and Engineering, Shanghai Engineering Research Center for Intelligent Instrument for Diagnosis and Therapy, Key Laboratory for Thin Film and Microfabrication of Ministry of Education, Department of Instrument Science and Engineering, School of Electronic Information and Electronical Engineering, 800 Dongchuan Road, Shanghai Jiao Tong University, Shanghai 200240, P. R. China.
3 Consolidated Research Institute for Advanced Science and Medical Care Waseda University, 513 Wasedatsurumaki-cho Shinjuku-ku, Tokyo 162-0041, Japan.
* Corresponding authors. E-mail: dxcui@sjtu.edu.cn; jiannihome@yahoo.com
Received: Mar. 28, 2017; Accepted: Mar. 29, 2017; Published: Mar. 31, 2017
Citation: Chenlu Li, Fangfang Xia, Kan Wang, Can Wang, Ping Xu, Hong Zhang, Jinping Wang, Asahi Toru, Jian Ni, and Daxiang Cui, Dendrimer-Modified Gold Nanorods as High Efficient Controlled Gene Delivery Release System under Near-Infrared Light Irradiation. Nano Biomed. Eng., 2017, 9(1): 82-95.
DOI: 10.5101/nbe.v9i1.p82-95.
Gold nanorod-based gene delivery system potentially represents a powerful nanotechnology for cancer therapy. Here we for the first time reported the use of polyamidoamine (PAMAM) dendrimer modified-gold nanorods (dGNR) as a high efficient gene delivery system for the targeted silencing of survivin via RNA interference for breast cancer therapy. Gold nanorods were functionalized with thiol-terminated polyamidoamine dendrimer; survivin shRNA plasmid was constructed and conjugated with dGNR. The resultant survivin-shRNA-GNR nanocomposites were incubated with human breast cancer MCF-7 cells, irradiated with 140 mJ/pulse of laser light of 1,064 nm for 15 s, and then continued to culture for 1 to 3 days. These cells were collected and analyzed by MTT, quantitative reverse transcription-PCR, Western blotting, fluorescent microscopy, and high-resolution transmission electron microscopy. 25 nude mice models with breast cancer were established; the nanocomposites of survivin-shRNA-dGNR were injected into the tumor tissues at gradually increased dose, and then were irradiated with 140 mJ/pulse of laser light of 1,064 nm for 15 s one time per week. The nude mice were raised for two months, and then were sacrificed. The tumor tissues were picked out, and their sizes were measured. Results showed that PAMAM dendrimer-functionalized gold nanorods were successfully synthesized; they could enter into MCF-7 cells within 30 min, release survivin shRNA plasmids with high efficiency, enhance the expression of transferred survivin genes in tumor cells under near-IR laser irradiation, and cause remarkable down-regulation of survivin gene and protein, inhibite cell growth in dose-and time-dependent means, and induce cell apoptosis. EGFP fluorescent signals in the tumor localization of nude mice. The tumor sizes in nude mice became smaller and smaller as the dose of the injected nanocomposites increased. In conclusion, PAMAM dendrimer-functionalized gold nanorods may be a high efficient gene delivery release system for survivin-shRNA vector under near-infrared light irradiation; the constructed survivin-shRNA-dGNR composites can effectively inhibit the growth of breast cancer cells, and have potential applications in breast cancer therapy and molecular imaging.
Key words: Gold nanorods; Polyamidoamine dendrimer; Survivin; RNA interference; Near-infrared laser irradiation
Introduction
Gold nanorod-based gene delivery system potentially represents a powerful nanotechnology for cancer therapy. Gold nanords (GNRs), which subordinate to the number of noble metal nanoparticles, are attracting considerable attention for their unique properties in photothermal therapy, biosensing, imagining and gene delivery for cancer [1-3]. GNRs are elongated gold nanoparticles with particular optical properties depending on their size and shape [4,5]. However, being completely different from spherical gold nanoparticles, GNRs split into two surface plasmon absorption bands. One is called transverse plasmon band, relating to the absorption and scattering of light along short axis of rods, located in the visible region at ca. 520 nm; the other is called longitudinal plasmon band, relating to the absorption and scattering of light along long axis of rods, located in the near-infrared region (NIR) of electromagnetic spectrum which is tunable by changing the aspect ratio of GNRs. It is well known that all the biologic tissues are relatively transparent in NIR where there are many exciting applications for photodynamic and photothermal therapy of cancer. Based on this, GNRs are extremely attractive candidates with essential properties as optical sensors for biological and medical applications. So far, there have been some reports closely associated with the use of gold nanorods as gene delivery system and controlled plasmid release [6-8]. Although GNRs exhibit the promising future, there are still some concerns closely associated with their biocompatibility. How to get rid of the toxic surfactant cetyltrimethylammonium bromide (CTAB) on the surface of gold nanorods and keep their good dispersion ability is still a challengeable problem in biomedical application. Dendrimers are a novel special class of organic molecules. They can take different functional groups through a series of chemical modifications, and their interior cavities can serve as storage areas for a lot of genes or drugs [9,10]. Dendrimers have been confirmed to be a good nonviral delivery vector because it has the advantages of safety, simplicity of use and ease of mass production. We have confirmed that the dendrimer can functionalize nanomaterials such as CNTs, quantum dots and magnetic nanoparticles, and markedly enhance their biocompatibilities and cellular uptakes of nanoparticles [11,12]. In this study, we fully used the advantages of GNRs and dendrimers, and selected survivin gene and breast cancer cell line MCF-7 cells and nude mice model with breast cancer as research targets. We constructed the pCMV-EGFP-U6-survivin-shRNA plasmid, and selected No. 5 generation of polyamidoamine (PAMAM) dendrimer-modified gold nanorods as the delivery system for survivin-shRNA plasmid. We investigated the delivery efficiency and biocompatibility of the dendrimer-functionalized gold nanorods, the survivin-shRNA controlled release ability in MCF-7 cells and tumor tissues under near-infrared laser irradiation, as well as the in vitro and in vivo biological effects of survivin-shRNA plasmids. The aim was to develope a safe, highly efficient, controlled gene delivery release system for breast cancer therapy.
Materials and Methods
Materials
Chloroauric acid (HAuCl4·3H2O), trisodium citrate, sodium borohydride (NaBH4) and ascorbic acid were obtained from Aldrich Company. Cetyltrimethylammonium bromide (CTAB) and Methyl mercaptoacetate (98%) were obtained from Sigma Company. All the glassware were cleaned by H2SO4 and rinsed with de-ionized water prior to experiments. Poly(amidoamine) (PAMAM) dendrimers with surface amine groups were stored in our lab. A human breast cancer cell line MCF-7 was obtained from ATCC Company. An RPMI 1640 medium containing 10% fetal calf serum (FCS) was from Gibco Company. Agarose was from Sigma Company. 3-(4,5-Dimethyl-2-thiazolyl) -2,5-diphenyl- 2H-tetrazolium bromide (MTT) was also obtained from Sigma. All enzymes were purchased from Promega Company.
Fabrication
Preparation of gold seed: 10 ml aqueous solution containing 2.5×10-4 M HAuCl4 and 2.5×10-4 M tri-sodium citrate was prepared in a conical flask. Next, 0.6 ml of ice-cold 0.1 M NaBH4 was added to the solution all at once while it was being stirred. The solution turned pink immediately after the NaBH4 was added, indicating particle formation. The particles in this solution were used as seeds within 2-5 h after preparation [13,14].
Preparation of 3.0 aspect ratio gold nanorods: In a clean test tube, 10 ml of growth solution, containing 2.5×10-4 M HAuCl4 and 0.1 M cetyltrimethylammonium bromide (CTAB), was mixed with 0.05 ml of 0.1 M freshly prepared ascorbic acid solution. Next, 0.025 ml of the 3.5 nm seed solution was added. There was no further stirring or agitation. Within 10 min, the solution color changed to reddish brown. After 48 h incubation, the solution contained 3.0 aspect ratio nanorods.
Preparation of thiol-terminated PAMAM dendrimer: The proper amount of methyl mercaptoacetate (NH2: SH=64:1) in water was added to 10 mg freshly prepared G5.0 amine-terminated PAMAM dendrimer dissolved in 20 ml water. The mixture was stirred at 50 ˚C for 9 h to complete the formation of the thiol-terminated dendrimer. The solvent was then removed under vacuum at 70 ˚C to obtain thick, honey-colored oil [15]. The chemical structure of thiol-modified PAMAM dendrimers is shown in Fig. 1.
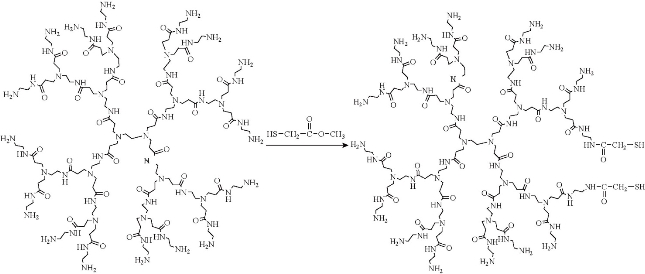
Fig. 1 Amine terminated No. 5 generation of PAMAM dendrimers reacted with methyl mercaptoacetate to form thiol-terminated PAMAM dendrimer.
Covalent binding of thiol-terminated dendrimer with gold nanorods
The gold nanorod solution described above and a 1% (wt%) thiol-terminated dendrimer aqueous solution were mixed at the ratio of [PAMAM]/[Au] = 10/1 (mol/mol) and incubated for 5 h at room temperature, to form stable aqueous solutions containing dendrimer-coated gold nanorods (dGNR). The resultant dGNR composites were centrifuged, and washed with distilled water for 3times, for further usage, as shown in Fig. 2.
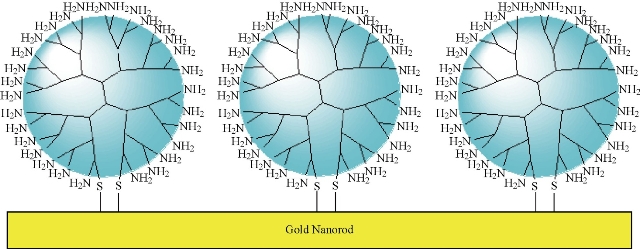
Fig. 2 Schematic representation of a nonviral gene delivery system based on a PAMAM dendrimer-coated gold nanorod: covalent binding of thiol-terminated PAMAM dendrimer on gold nanorod surface to form dendrimer coated gold nanorods (dGNR).
Characterization of dendrimer-coated gold nanorods by AFM, TGA, UV-Vis spectroscopy, and HR-TEM
High-resolution transmission electron microscopy (HR-TEM, Hitachi H-700H) was used to confirm particle size and observe dendrimer coating. Thermogravimetric analysis (TGA) measurement was performed by a TGA 2850 thermogravimetric analyzer (TA Instruments) under N2 in the temperature range of 30–700 ˚C with an increasing rate of 5 ˚C/min. Atomic force microscopy (AFM) imaging was performed by a Nanoscope III (Digital Instruments/Veeco Metrology Group, USA). AFM images were obtained in the tapping mode with standard Si/N tips. UV–Vis (ultraviolet–visible spectroscopy) spectra were measured at 20 ˚C with a UNICAM UV 300 spectrometer (Thermo Spectronic, U.S.A.) equipped with a 10 mm quartz cell. The 400–1100 nm wavelength region was scanned, since it includes the absorbance of the gold nanorods.
Construction of surviving-shRNA expressing plasmid
The pEGFP-U6-shRNA vector [16] was presented as a gift by Professor Beate Brand-Saberi from Freiburg University, Germany. The survivin-shRNA vectors were constructed as follows.
(1) The selected target sequence in the survivin gene (accession number: NM_001168) was from nucleotide 51 to 70. The target sequence was selected with the help of the Whitehead siRNA selection program (Bioinformatics and Research Computing at Whitehead Institute). A Blast search against the NCBI database revealed no significant homology of the targeted sequence to other genes. As shown in Fig. 3, as a survivin shRNA DNA template, one oligo sequence subsequently should include a sense sequence (5’-CACCG CATCT CTACA TTCAA GAATT CAAGA GATTC TTGAA TGTAG AGATG CTTTT TTG-3’), a loop sequence (5’-TTCAAGAGA-3’), and an antisense sequence (5’-GATCC AAAAA AGCAT CTCTA CATTC AAGAA TCTCT TGAAT TCTTG AATGT AGAGA TGC -3’). In order to achieve the ligation of the vectors, two enzyme restriction sequences for BamHI (at the 5’ end) and Hind III (at the 3’end) must locate at both ends. For the purpose of later convenient analysis of the DNA plasmid constructs, an EcoR I restriction sequence should be added before the Hind III sequence.
(2) All oligonucleotides were synthesized by the Ambion Company and purified using polyacrylamide gel. They were dissolved in an annealing buffer (10 mM Tris-Cl (pH 8.0), 50 mM NaCl, 1 mM EDTA) and annealed by heating to and stepwise cooling as follows: 95 ˚C, 4 min; 90 ˚C, 4 min; 85 ˚C, 4 min; 80 ˚C, 4 min; 75 ˚C, 4 min; 70 ˚C, 4 min; 65 ˚C, 4 min; 60 ˚C, 4 min; 55 ˚C, 4 min; 50 ˚C, 4 min; 45 ˚C, 4 min; 40 ˚C, 4 min; 35 ˚C, 4 min; 30 ˚C, 4 min; and 25 ˚C, 4 min. The oligonucleotide-duplexes codings for survivin and scrambled shRNAs were ligated to BamH I- and Hind III-cleaved pEGFP-U6-shRNA vectors to generate a pEGFP-U6-shRNA-survivin vector respectively. After that, the insertion sequences were confirmed by sequencing, and the blank control vectors were constructed according to the method described in reference [17].
Binding of pEGFP-U6-survivin-shRNA vector with dGNR
The pH of the solution was 7.4. A suspension of the dGNR at a concentration of 0.025 mg/ml was mixed with 1 μM pEGFP-U6-survivin-shRNA vectors for 2 h at room temperature prior to characterization or cellular incubation for intercellular uptake. The pEGFP-U6-survivin-shRNA vectors were adsorbed nonspecifically onto the dGNR surface via electrostatic attraction.
Agarose gel electrophoresis
0.7% agarose gel electrophoresis was used to assess the formation of the dGN-pEGFP-U6-survivin-shRNA vector complex. The nanocomplexes were added to the wells of the ethidium-bromide agarose gel; the resulting gel was processed at 60 V for 20 min and then imaged using a Chemi-Doc system (Bio-Rad, Hercules, CA).
Cell culture and observation by fluorescent microscopy and HR-TEM
Human breast cancer MCF-7 cells were cultured at 37 ˚C in a humidified 5% CO2 and 95% air atmosphere in DMEM containing 1×105 mU/ml of penicillin and 0.1 mg/ml of streptomycin supplemented with 10% (v/v) FCS. The MCF-7 cells were collected, seeded into 12 well plates, and cultured for 24 h as the experiment cells. Next, 100 μL dGN-pEGFP-U6-survivin-shRNA vector complexes were seeded into 6 well plates. Then, these blank control vectors and the dGNR composites were seeded into other 6 well plates as the control group. These cells were irradiated with 140 mJ/pulse of laser light of 1,064 nm for 15 s; the control group was set without near-IR irriadiation. Finally, all the cells were cultured at 37 ˚C for 30 min, 40 min, 1 h, 24 h, 48 h, and 72 h. After culturing, these cells were observed by fluorescent microscopy. Then, the MCF-7 cells were collected and embedded into paraffin and made into TEM specimens, which then were observed via HR-TEM.
Cell viability and proliferation assays
The MCF-7 cells were cultured in a DMEM medium supplemented with 10% FCS and 1% penicillin-streptomycin at 37 ˚C for 48 h. The MCF-7 cells were collected and added into 24 well plates at the concentration of 5000 cells/well then; they were allowed to culture for 24 h. Next, the 0.025 mg/ml dGNR-pEGFP-U6-survivin-shRNA vector nanocomplexes were added into the 24 well plates; at the same time, the control experiments with the dGNR-pEGFP-U6-shRNA blank vector or dGNR were set up and continued to culture for 24 h, 48 h, 72 h. MTT (5 mg/ml) was prepared in PBS and 20 μL was added to each well; the cells were incubated for 4 h at 37 ˚C; the medium was removed; 200 μL dimethyl sulfoxide (DMSO) was added to each well. The optical density (OD) of each well was determined to be 515 nm, according to the microplate reader. The cell viability was calculated by the following formula: cell viability (%) = OD(optical density) of the treated cells/ OD of the non-treated cells [18].
Semi-quantitative RT-PCR analysis
Cellular total RNAs were extracted out by using a Total RNA Extraction Kit according to the manufacturer’s instructions. Single-stranded cDNA was synthesized with oligo(dT) primer in a 20 l reaction from 5g of total RNA using the SuperScript Preamplification System for First Strand cDNA Synthesis (Promega Company, USA) and diluted up to 80 l. The PCR primers for the survivin gene were as follows: forward primer: 5’-GTG AAT TTT TGA AAC TGG ACA G-3’; reverse primer: 5’-CCT TTC CTA AGA CAT TGC TAA G- 3’; amplification fragment: 240 bp; β-actin (internal control) forward primer: 5’-TCC TGT GGC ATC CAC GAA ACT-3’; β-actin reverse primer: 5’-GAA GCA TTT GCG GTG GAC GAT-3’; products length: 180bp. PCR was then performed with 1l of cDNA for 1 cycle at 94˚C for 2 min, followed by 30 cycles, each at 94 ˚C for 30 s, then 60 ˚C for 30 s, and then 72 ˚C for 2 min using gene-specific primers and Taq polymerase. PCR products were analyzed by 1% agarose gel electrophoresis with ethidium bromide, and -actin was used as the internal control to confirm equal amounts of the templates [19].
Western blot analysis
MCF-7 cells were lysed in a protein lysis buffer (50mM Tris pH 7.4, 150mM NaCl, 1mM EDTA, 1mM EGTA, 5% 2-mercaptoethanol, 1% NP-40, 0.25% sodium deoxycholate, 5g/ml leupeptin, 5g/ml aprotinin, 10 g/ml soybean trypsin inhibitor, and 0.2mM phenylmethyl sulfonylfluoride). Protein concentrations were determined using the Bradford method. Equal amounts of sample lysate were separated by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). The membrane was blocked with 0.1% BSA in a TBST buffer (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.1% Tween-20) and incubated overnight at 4 ˚C with a rabbit anti-human polyclonal antibody against survivin. Subsequently, the membrane was washed with TBST buffer and incubated with a horseradish peroxidase-conjugated secondary antibody. Enhanced chemiluminescence kits were used (Amersham ECL kits) to enhance the fluorescence of the bands. β-actin was used as a negative control [20].
Preparation of Nude mice models with breast cancer
20 nude mice with identical weights were purchased from animal experimental center in Shanghai JiaoTong University. The experiment was approved by animal experiment committee. These mice were divided into 4 groups. Their backs were injected with 500 l MCF-7 cells by hydromeic, and raised for two weeks. The tumors on their backs can be observed. They were then injected with 25 mol GNRs, 25 mol dGNRs, survivin-shRNA-dGNRs (25 mol, 50 mol, 75 mol) respectively, irradiated with 1,046 nm laser for 15 s one time per week, and continued to culture for eight weeks. They were taken pictures by micro-pet, and then were sacrificed. Their tumors were picked up, of which the sizes were measured.
Statistical analysis
Results were calculated as the mean ±SE of the triplicate experiments. The significance of the results was statistically analyzed by a one-way analysis of variance (ANOVA) with Tukey’s multiple comparison for pairwise comparison. Statistical significance was set at P < 0.05.
Results
Characterization of dGNRs
Fig. 3 shows the results of G5.0 PAMAM dendrimer functionalized gold nanorods characterized by HR-TEM and AFM. As shown in Fig. 3(a), synthesized gold nanorods were 50 nm in length, and 15 nm in width. Fig. 3(b) shows there was a dendrimer layer covering on the surface of each gold nanorod. Fig. 3(c) shows that a tree-shapped dendrimer layer covered on the surface of the gold nanorods. As shown in Fig. 3(d), gold nanorods had two absorption bands, one at a shorter wavelength (534 nm) and the other at a longer wavelength (851 nm). After the nanorods were coated with PAMAM dendrimer, the two bands shifted to higher wavelengths, at 542 and 871 nm respectively. The dendrimer coating layer on the surface of GNRs were further confirmed by TGA analysis as shown in Fig. 3(e). The grafted dendrimer content (%) of the dGNR was calculated by TGA from the weight loss between 200 ˚C and 800 ˚C. For comparison, the TGA curve of gold nanorods without dendrimer coating is also shown. In the case of the gold nanorods, between 200 ˚C and 800 ˚C there was a continuous but not very obvious decrease (~ 4%) in weight, which is a typical weight loss for the CTAB layer on the surface of gold nanorods. The quantity of dendrimer attached to the gold nanorods is also shown in Fig. 1(e), there was a weight loss of ~ 17% for the dGNR. All of these reveal the existence of G5.0 PAMAM dendrimer on the surface of gold nanorods.
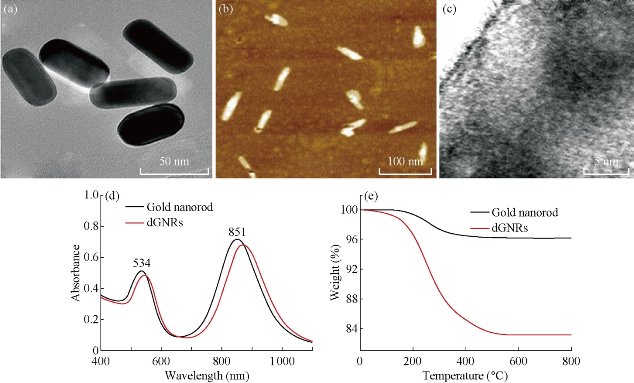
Fig. 3 HR-TEM and AFM images of gold nanorods with and without modified dendrimer. (a) HR-TEM image of gold nanorods without dendrimer coating; (b).AFM image of gold nanorods with dendrimer coating; (c) HR-TEM image of individual dendrimer-coated gold nanorod (dGN); (d).UV–Vis absorbance spectra of GNRs before and after dendrimer modification; (e) TGA curves of dGNRs.
Electrophoretic shift assay and zeta potential analysis
Fig. 4(a) shows the electrophoretic shift assay result of pCMV-EGFP-U6-survivin-shRNA plasmid and the blank control pCMV-EGFP-U6-shRNA plasmid digested with EcoR I. Compared with the control empty vector. The constructed pCMV-EGFP-U6-survivin-shRNA plasmid had a 156 bp fragment, which matched with the inserted survivin fragment; therefore, the pCMV-EGFP-U6-survivin shRNA plasmids (survivin-shRNA vector) were successfully constructed. Fig. 4(b) shows the electrophoretic shift assay result of survivin-shRNA vector in the absence and presence of dGNR, which indicates that the amine-terminated dMGR composites could bind with survivin-shRNA vector and formed stable survivin-shRNA-dGNR composites. Fig. 4(c) shows zeta potential analysis result of dGNR-survivin-shRNA composites and dGNR. After G5.0 dendrimer modified GNR, zeta potential reached +30 mV at pH 7.0, which was due to the increasing positive charges of –NH3+ on the surface of gold nanorods. The dGNR with positive charge could bind with survivin-shRNA vector with negative charge via electrostatic attraction. G5.0 dGNRs absorbed the biggest amount of survivin-shRNA vectors. Positively charged survivin-shRNA-dGNRs would be easily attached to negatively charged cell membrane to improve the endocytosis. More positive charges on survivin-shRNA-dGNRs will enhance the gene delivery efficiency.
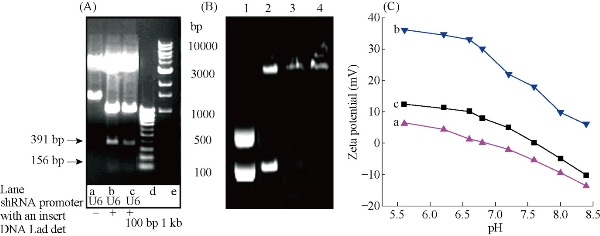
Fig. 4 Electrophoresis images: (A) Enzyme restriction analysis of the constructed shRNA plasmids with EcoR I. The plasmid with insert produced three bands; in contrast, the normal control plasmid with no insert had only two bands. (B) 0.7 % agarose electrophoresis mobility retardation assays: lane1: CMV-EGFP-U6-shRNA-survivin plasmids; lane2: mixture of pCMV-EGFP-U6-shRNA-survivin plasmids and gold nanorods; lane 3: dGNR-pCMV-EGFP-U6- survivin-shRNA vector composites; lane 4: dGNR-pCMV-EGFP-U6-blank –shRNA vector. (C) Zeta potentials of (a) CTAB-gold nanorods; (b) G5-dendrimer modified gold nanorods; (c) survivin-shRNA vector-dGNR composites.
Cell viability and proliferation inhibition assays
As shown in Fig. 5(a), G5.0 dGNRs could inhibit the growth of breast cancer cells with the inhibition rate of 6%, indicating that the dGNRs were very low toxic to breast cancer cells. Under identical condition, survivn-shRNA-dGNR composites had the inhibition rate of 60%; the control empty vector-dGNR composites only had the inhibition rate of 7%; there existed statistical difference between two groups (P < 0.01). This result represents that survivn-shRNA-dGNR composites can markedly inhibit the growth of breast cancer cells. As shown in Fig. 5(c), dGNR-survivin-shRNA composites inhibited the growth of tumor cells in dose-dependent means.
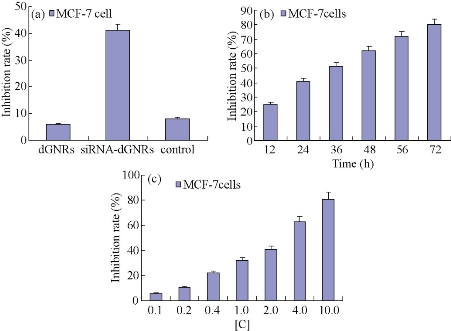
Fig. 5 Growth inhibition curves of MCF-7 cells. (a) dGNRs, shRNA-dGNRs and control at 24 h; (b) survivin shRNA-dGNRs composites at different time; (c) shRNA-dGNRs composites at different concentration. [C] = 0.025 mg/ml dGNRs + 2 μM survivin-shRNA vectors, control: 0.025 mg/ml dGNRs + 2 μM blank pCMV-EGFP-U6-shRNA vectors.
Semiquantitative reverse transcription-PCR and Western blot analysis
As shown in Fig. 6, as the amount of survivin-shRNA GNR composites increased, the expression levels of gene and protein of survivin in breast cancer cells exhibited gradually down-regulated expression compared with the control group; there existed a statistical difference between survivin-dGNR composites and control composites (P < 0.01). The result demonstrates that No. 5 generation of dendrimer-modified gold nanorods can be used as gene delivery system for cellular-targeted silencing of survivin gene by RNA interference. The result also shows that the design and construction of survivin-shRNA vector was very successful.
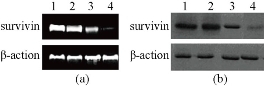
Fig. 6 QRT-PCR and Western blotting results showing the effect of dGN-pCMV-EGFP-U6-shRNA-survivin plasmid composites on the expression of survivin gene and protein in MCF-7 cells. (a) QRT-PCR analysis of survivin or β - actin (internal control) mRNA expression performed 48 h after transfection; (b) Western blot analysis of survivin or β-actin (internal control) protein expression performed 48 h after transfection; (lane 1) control experiments without adding dGNR-pCMV-EGFP-U6-shRNA-survivin plasmid composites; (lane 2) adding dGNR only; (lane 3) adding mixture of dendrimer-pCMV-EGFP-U6-shRNA-survivin plasmids; (lane 4) adding dGNR- pCMV-EGFP-U6-shRNA-survivin plasmid complex.
Morphologic observation of breast cancer cells with dGNR-survivn-shRNA vectors
As shown in Fig. 7, MCF-7 cells with dGNRs or dGNR-blank shRNA vectors grew very well; as the dose of dGNR-survivin-shRNA composites in culture medium and culture times increased gradually, partial tumor cells gradually became round and suspended, and finally exhibited apoptosis or death characterization. To further confirm that the dGNR-survivin-shRNA nanocompsoites were indeed internalized by the MCF-7 cells, HR-TEM images were taken on MCF-7 cells cultured with dGNR-survivin-shRNA composites for 30 min, 40 min and 1 h. As shown in Fig. 8(a), dGNR-survivin-shRNA composites can be observed to locate in cytoplasm while incubated with MCF-7 cells for 30 min, which fully shows that dGNR-survivin-shRNA composites could enter into tumor cells in high efficiency. In Fig. 8(a)-(c) & (f), HR-TEM images showed that a large number of dGNR-survivin-shRNA composites were located in the cytoplasm, and few composites were in the nuclei. In Fig. 8(d)-(f), HR-TEM images showed that dGNR-survivin-shRNA composites in the in vitro solution and in vivo tumor tissues could become pieces under the near-IR irradiation, which enhanced the survivin-shRNA vectors release from the composites, and finally enhanced the inhibition effects of survivin-shRNA in the tumor tissues. As shown in Fig. 8(g) & (h), the MCF-7 cells transferred with GNR-survivin-shRNA composites without near-IR irradiation had 60% or so positive signal; the MCF-7 cells transferred with GNR-survivin-shRNA composites with near-IR irradiation had almost 90% or so positive signal, which indicates that near-IR irradiation may markedly enhance the positive expression of the transferred survivin genes in MCF-7 cells.
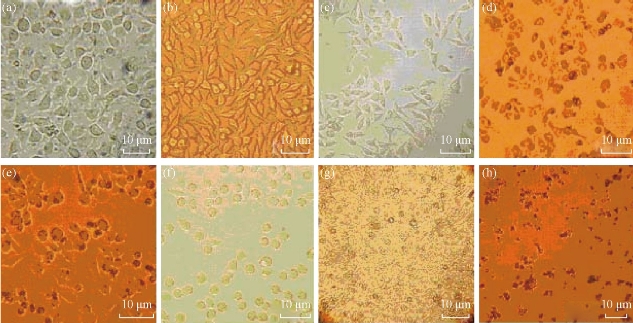
Fig. 7 Observation of cellular morphology by optical microscopy after 24 h ~ 72 h incubation. (a) MCF-7 cells with dGNRs for 48 h; (b) normal culture MCF-7 cells for 48 h; (c)-(e) MCF-7 cells with 25 g/ml, 50 g/ml, 75 g/ml dGNR-survivin-shRNA composites for 24 h, respectively; (f)-(h) 25 g/ml dGNR-survivin-shRNA composites for 24 h, 48 h and 72 h, respectively.
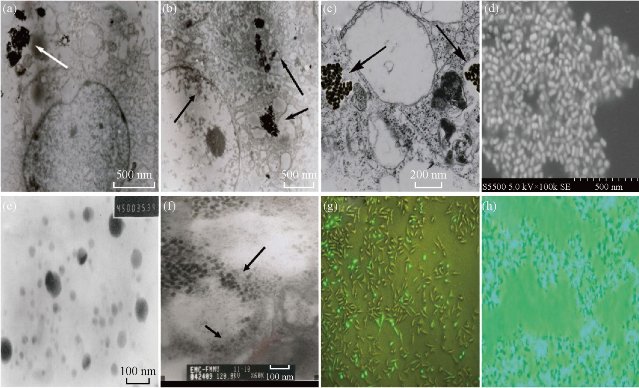
Fig. 8 Images of dGNR-survivin-shRNA composites in in vitro and in vivo cells.(a)-(c) HR-TEM images of dGNR-survivin-shRNA complexes (arrow) in cytoplasm of MCF-7 cells incubated with cells for 30 min, 40 min and 1 h, respectively; (d) SEM image of dGNR-survivin-shRNA composites; (e) HR-TEM image of dGNR-survivin-shRNA composites in solution after the near-IR irradiation for 15 s; (f) HR-TEM image of pieces of dGNR-survivin-shRNA composites in tumor tissues after the near-IR irradiation for 15 s, with the arrow indicating the gold nanoparticles inside the tumor tissues; (g) fluorescent microscopy imaging of MCF-7 cells transferred with GNR-survivin-shRNA composites without near-IR irriadiation for 24 h; (h) fluorescent microscopy imaging of MCF-7 cells transferred with GNR-survivin-shRNA composites after near-IR irradiation for 24 h (bar: 5 m).
Effects of dGNR-survivin-shRNA composites on sizes of tumors of nude mice
25 nude mice models with breast cancer were established successfully. In the whole course of experiments, nude mice grew very well; no one nude mouse died. As shown in Fig. 9(a), EGFP fluorescent signals can be observed in the tumor position of nude mice. The tumor sizes in nude mice became smaller and smaller as the injected dose of dGNR-survivin-shRNA nanocomposites increased, as shown in Fig. 9(b) & (c), which indicates that survivin-shRNA vectors could be released from the dGNR-survivin-shRNA composites in the in vivo microenvironment, exhibited strong positive expression in tumor tissues, and could inhibit the growth of tumor cells, whereas the control groups including the dGNR group and blank shRNA–dGNR group almost exhibited no inhibition of tumor tissues. All these results fully demonstrate that dGNRs may be a safer, and highly efficient gene delivery system; the survivin-shRNA vectors can be effectively used for breast cancer therapy.
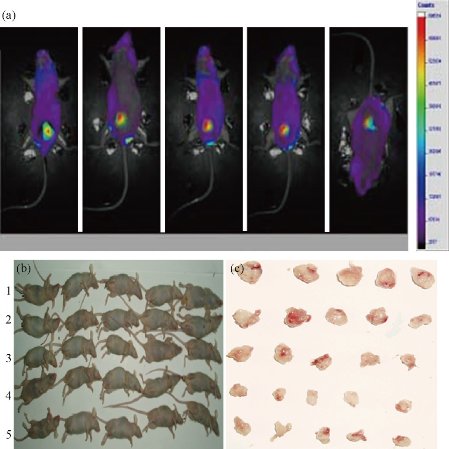
Fig. 9 Nude mice model loaded with breast cancer. (a) The imaging of tumors in the mice with dGNR-survivin-shRNA vector by micro-pet; (b) the nude mice were divided into 5 experiment groups, each group had 5 mice, No. 1 group: treated with dGNR, No. 2 group: treated with 25 mol dGNR-blank-shRNA composites, No. 3, No. 4 and No. 5 groups: respectively treated with 25 mol, 50 mol, 75 mol dGNR-survivin-shRNA composites. (c) tumor tissues cut from nude mice matched with experiment groups.
Discussion
RNA interference (RNAi) has become a powerful tool for the study of gene functions, and also is considered to be very promising for the gene therapy of cancer [21-26]. Some siRNA-expressing plasmids such as shRNA vectors, etc. have been constructed to provide a large amount of high efficient siRNAs for cells [27,28]. However, there exist some problems that need to be solved immediately, such as low transfection efficiency and fast degradation of the delivered genes asODN, siRNA, etc. by enzymes in vivo [29,30]. Virus based delivery vectors have higher transfection efficiency, but have the potential risk of inducing gene mutation and tumor [31]. Developing nonviral vectors should be a better alternative choice to solve the present problem. With the advancement of nanotechnology, lots of new nanomaterials with specifical function are being continuously fabricated, which makes it possible to find one kind of novel nanomaterial to act as a good delivery system for gene therapy[32]. Gold nanorods and dendrimer molecules have been synthesized controllably, and own potential applications in biological medicine and bioengineering [33,34]. Particularly, gold nanorods have been confirmed to be able to conjugate with biomolecules such as antibody or peptides by covalent bond or thoil bond, and enter into tumor cells by endocytosis pathway. Gold nanorods inside tumor cells or tissues can be observed by SERS (surface-enhanced Raman scattering), and can be controlled by near-infrared irradiation as well. Therefore, gold nanorods should be the best choice for gene or drug delivery [35,36]. Dendrimers represent a relatively new class of well defined monodisperse macromolecules with striking features, such as controlled structure, globular shape and a high density of “tunable” surface functional groups in their periphery. A variety of dendrimers have been synthesized and tailored for diverse applications. Various strategies have been devised to load dendrimers with drug molecules, genetic materials, targeting agents, dyes and imaging agents, by encapsulation, complexation or conjugation. Although the investigation of cellular delivery of gene and drugs using dendrimers is at an early stage, these nanomaterials offer several advantages when compared to liposomes and microparticles. Their multiple surface functional groups can be easily modified to potentially attach a large number of drug and gene molecules, ligands and antibodies, making them ideal vehicles for targeted gene delivery [37,38]. Herein, we selected gold nanorods and dendrimer molecules as raw materials, fabricated dendrimer-GNR composites, and investigated their across membrance efficiency and biological effects, with the aim of developing one novel kind of safe, high-efficient and nonvirus gene delivery system for cancer therapy. It is well known that survivin protein is one stronger inhibitor of apoptosis protein family due to its complex mode of antiapoptotic action and its additional function in the control of cell division. Survivin gene was found to be overexpressed in embryo tissues, malignant tumours and have no expression in normal tissue [39,40]. Therefore, survivin gene was an ideal target for cancer therapy. We constructed the survivin shRNA vectors and selected the blank shRNA vector as negative control. G5-dendrimer-modified GNR composites were fabricated, and characterized by HR-TEM, UV-spectroscopy, TGA and AFM. The dendrimer-modified GNRs were mixed with survivin shRNA vectors in the charge ratio of 3:1, under this condition, dendrimer-GNRs can absorb the maximum of anti-survivin shRNA vectors. According to the quality of dendrimer, we can account out the charges of dendrimer surface, therefore, the maximum of survivin shRNA vectors combined with dendrimer can be speculated out, this is one kind of approximate quantitative method. Electrophoretic shift assay and zeta potential analysis also confirmed that survivin shRNA vectors and blank shRNA vector really conjugated with dendrimer-GNRs. In order to investigate the efficiency of across cell membrance of dGNR-survivin-shRNA, we used optical microscopy to observe morphological changes of MCF-7 cells incubated with dGNR-survivin-shRNA composites, and used HR-TEM to observe the specimens of MCF-7 cells incubated with dGNR-survivin-shRNA composites for 30 min, 40 min and 1h. Results showed that dGNR-survivin-shRNA composites could enter into tumor cells within 30 min of incubation, the dGNR-survivin-shRNA composites were further confirmed to locate in the cytoplasm and nuclear by HR-TEM, similar to other repors [6]. According to previous reports, the peptide-conjugated GNR could be observed at 2 h or so of incubation with tumor cells; the CTAB-conjugated GNR’ delivery efficiency was very low (10% or so), compared with our results; there was statistical difference between two groups (P < 0.01), The result indicates that dendrimer can improve the across membrance efficiency of gold nanorods, which suggests that dendrimer-modified gold nanorods is likely one kind of good gene delivery system for breast cancer therapy. Previous reports [41, 42] showed that as a gold cube of nanoscale size became bigger, the absorption of infrared rays became stronger. This phenomenon can be used to enhance the effects of treatment by local in vitro or in vivo irradiation by infrared rays. Our results show that gold nanorods have a controllable absorbance band at ~ 850 nm; thus, gold nanorods coated with dendrimers can be used to track the position of pCMV-EGFP-U6-shRNA-survivin plasmid in cancer cells; more importantly, they also can be used to enhance therapy effects by local near–IR irradiation. As we observed, near–IR irradiation could enhance the positive expression ratio of transferred survivin genes in MCF-7 cells. Conversely, gold nanoparticles did not have a controllable absorbance band. Rosi, et al [43] reported that gold nanoparticles were not toxic to cancer cells, and could conjugate with genes or drugs and then deliver them into cancer cells for cancer therapy. However, one dendrimer-modified gold nanorod can deliver more genes or drugs than one gold nanoparticle; therefore, dendrimer-modified gold nanorods may have more delivery advantages than currently reported systems such as magnetic nanoparticles, quantum dots, gold nanoparticles, etc. Our experiment also shows that dendrimer-modified gold nanorods are almost nontoxic to cells. Therefore, so far the dendrimer-modified gold nanorods may be one highly efficient delivery system with good biocompatibility. In order to investigate the effects of survivin-shRNA vectors within the dGNR-survivin-shRNA composites on MCF-7 cells, we observed that the tumor cells cultured with dGNR-survivin-shRNA composites gradually became round and suspended, and that apoptosis appeared as the culture time increased. Conversely, the cells in the control group with dGNR-blank-shRNA composites and with dGNRs grew very well. These results showed that survivin-shRNA vectors in dGNR-survivin-shRNA composites took effects against cell proliferation. In order to further confirm this point, semiquantitative RT-PCR were used to detect the expression levels of survivin gene in tumor cells. Results showed that survivin gene in MCF-7 cells with anti-survivin-shRNA vectors gradually exhibited down-regulation expression as the dose of survivin-shRNA-dGNRs and culture time increased. Western blot analysis also confirmed that survivin protein in MCF-7 cells also exhibited down-regulation expression as the amount of dGNR-survivin-shRNA composites and culture time increased. Conversely, the cells in the control group exhibited no marked change in the expression of survivin gene and protein. These results fully showed that survivin-shRNA vectors of dGNR-survivin-shRNA composites could bind with the start site of survivin mRNA, block the translation of survivn mRNA into survivin protein, and finally down-regulate the expression of survivin gene and protein. These results also showed that dGNRs did not affect the function of survivin shRNA vectors in tumour cells. The survivin-shRNA vectors could escape from the survivin-dGNRs nanocomposites under the cell in vivo environment, and then survivin-shRNA vectors themselves bound with the start site of survivin mRNA and block translation of survivin mRNA into survivin protein. The dGNRs may be expelled out of cells. The concrete course and mechanism need further clarifications. As the survivin-shRNA partner, the dGNR may prevent the fast degradation of survivin-shRNA by enzymes in vivo cells, and extend the live time of survivin-shRNA vector inside cells. Because dendrimer-GNRs can change the cellular micro-environment within a short time, absorb some ions, and change the pH of the environment, thus protecting survivin-shRNA vectors from degradation as caused by in vivo enzymes. Lots of research reports showed that siRNA take effects within 2 days after entering into tumor cells. As the time increased, the function of siRNA disappeared gradually. In this study, we observed that as the incubation day increased, the cell growth inhibition became more and more marked. Therefore, we suppose that dendrimer-GNRs may be helpful to protect survivin-shRNA vectors from fast degradation by enzymes in vivo. In order to further investigate the biological effects of dGNR-survivin-shRNA composites, we prepared nude mice models with breast cancer, and directly injected the nanocomposites of dGNR-survivin-shRNA into tumor tissues of nude mice. We observed that these nude mice did not die within 2 months. We also observed that EGFP fluorescent signals in the tumor location of nude mice and tumor sizes in the nude mice became smaller and smaller as the dose of dGNR-survivin-shRNA composites increased. According to the above results, we suggest a possible model of interaction between dGNR-survivin-shRNA and tumor cells. The survivin-shRNA vectors combined with dendrimer modified GNRs via electrostatic means. The resultant dGNR-survivin-shRNA nanocomposites had positive charges on their surface, and were easily attached to the surface of tumor cells, inducing the internalization of dGNR-survivin-shRNA composites into tumor cells by endocytosis means. Under the near-infrared irradiation, the gold nanorods inside cells or tumor tissues could turn into sphere pieces. Under the action of lysosome enzymes and ions in cells, survivin-shRNA vectors escaped from dGNR-survivin-shRNA composites, entered into ribosome, bound the start sites of survivin mRNA, blocked the translation of survivin mRNA into survivin protein, eliminated the function of survivin protein against apoptosis, and induced cell apoptosis. The dGNRs may be expelled out of in vivo cells. The concrete mechanism needs further clarifications. In conclusion, dendrimer conjugated gold nanorods were successfully fabricated, and were nontoxic to tumor cells. These resulting dendrimer-gold nanorods composites could bind with survivin-shRNA vectors via static-electronic means. G5-dendrimer–modified gold nanorods exhibited the maximum amount of combined survivin-shRNA vectors. Dendrimer coating layer markedly improved the dispersion and biocompatibility of gold nanorods. The dGNR-survivin-shRNA composites could cross the cell membrane, enter into tumor cells within 30 min, and inhibit the growth of tumor cells. The dendrimer-GNRs almost did not affect the growth of tumor cells, were helpful to protect survivin-shRNA vectors from degradation by enzymes inside cells, and almost did not affect the function of survivin-shRNA vectors in cells. The survivin-shRNA-dendrimer–conjugated gold nanorods exhibited the maximum inhibition against tumor cell growth. The dGNRs had a controllable absorbance band at ~ 850 nm, which can be used to monitor their position in cancer cells and also can be used to enhance the treatment by local infrared irradiation. Although the concrete metabolism of shRNA-dGNRs inside cells was not well clarified, the dendrimer-conjugated gold nanorods may be one kind of safe and highly efficient gene delivery system, and have potential application in breast cancer therapy and molecular imaging.
Acknowledgements
This work was supported by China National 973 Project (No. 2015CB931802 and No. 2010CB933901), China National 863 Project (No. 2007AA022003 and 2014AA020700), National Natural Scientific Fund (No. 30471599 and No.30672147), , and Shanghai Fund of Science and Technology (No.054119527 and 0752nm024).
References
1 X.H. Huang, P.K. Jain, I.H. El-Sayed, et al., Gold nanoparticles: interesting optical properties and recent applications in cancer diagnostic and therapy. Nanomedicine, 2007, 2: 681-693.
2 C.D. Chen, S.F. Cheng, L.K. Chau, et al., Sensing capacity of the localized surface plasmon resonance of gold nanorods. Biosens Bioelectron, 2007, 22: 926-932.
3 A.K. Salem, P.C. Searson, and K.W. Leong, Multifunctional nanorods for gene delivery. Nat Mater, 2003, 2: 668-671.
4 N.R. Jana, L. Gearheart, and C.J. Murphy, Wet chemical synthesis of high aspect ratio cylindrical gold nanorods. J Phys Chem B, 2001, 105: 4065-4067.
5 J. Perez-Juste, I. Pastoriza-Santos, L.M. Liz-Marzan, et al., Gold nanorods: synthesis, characterization and applications. Coordin Chem Rev., 2005, 249: 1870-1901.
6 A.K. Oyelere, P.C. Chen, X.H. Huang, et al., Peptide-conjugated gold nanorods for near targeting. Bioconjugate Chem, 2007, 18: 1490-1497.
7 Y.F. Huang, H.T. Chang, and W.H. Tan, Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal. Chem, 2008, 80: 567-572.
8 J.W. Lee, B.K. Kim, H.J. Kim, et al., Convergent synthesis of symmetrical and unsymmetrical PAMAM dendrimers. Macromolecules, 2006, 39: 2418-2422.
9 S. Hong, P.R. Leroueil, E.K. Janus, et al., Interaction of polycationic polymers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem, 2006, 17: 728-734.
10 I.J. Majoros, A. Myc, T. Thomas, et al., PAMAM dendrimer-based multifunctional conjugate for cancer therapy: synthesis, characterization, and functionality. Biomacromolecules, 2006, 7: 572-579.
11 B. Pan, D. Cui, Y. Shen, et al.,Using dendrimer modified magnetite nanoparticles as gene delivery system for cancer therapy. Cancer Research, 2007, 67: 8156-8163.
12 B. Pan, D. Cui, P. Xu, et al., Cellular uptake enhancement of polyamidoamine dendrimer modified single walled carbon nanotubes. Journal of Biomedical & Pharmaceutical Engineering, 2007, 11: 1-4.
13 B. Pan, D. Cui, C.S. Ozkan, et al., DNA-templated ordered array of gold nanorods in 1d and 2d. J Phys Chem C, 2007, 111(34): 12572-12576.
14 B. Pan, L. Ao, F. Gao, et al., End-to-end self-assembly and colorimetric characterization of gold nanorods and nanospheres via oligonucleotide hybridization. Nanotechnology, 2005, 16: 1776–1780.
15 T. Sara, B. Esmaeil, Modification of carbon nanotubes as an effective solution for cancer therapy. Nano Biomed. Eng., 2016, 8(3): 144-160.
16 F. Dai, F.Yusuf, G.H. Farjah, et al., RNAi-induced targeted silencing of developmental control genes during chicken embryogenesis. Dev Biol, 2005, 285: 80-90.
17 R. Bernards, T.R. Brummelkamp, R.L. Beijersbergen, et al., shRNA libraries and their use in cancer genetics. Nature Methods, 2006, 3 (9): 701-706.
18 D. Cui, F. Tian, M. Wang, et al., Effects of single walled carbon nanotubes on HEK293 cells. Toxicol Lett, 2005, 115: 73-85.
19 S. Derzelle, F. Dilasser, A robotic DNA purification protocol and real-time PCR for the detection of Enterobacter sakazakii in powdered infant formulae. BMC Microbiol, 2006, 6: 100 -104.
20 D. Cui, G. Jin, T. Gao, et al., Characterization of BRCAA1 and its novel antigen epitope identification. Cancer Epidem. Biomar. Prev, 2004, 13: 1136-1145..
21 A.J. Bridge, S. Pebernard, A. Ducraux, et al., Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genetics, 2003, 34(3): 263-264.
22 D. Shirane, K. Sugao, S. Namiki, et al., Enzymatic production of RNAi libraries from cDNAs. Nature Genetics, 2004, 36(2): 190-196.
23 M. Miyagishi, H. Sumimoto, H. Miyoshi, et al., Optimization of an siRNA-expression system with an improved hairpin and its significant suppressive effects in mammalian cells. J Gene Med, 2004, 6: 715–723.
24 T. Huynh, S. Walchli, and M. Sioud. Transcriptional targeting of small interfering RNAs into cancer cells. Biochemical and Biophysical Research Communications, 2006, 350: 854–859.
25 A.M.Adams, S.L.Pratt,, S.L.Stice, Knockdown of the dnmt1s transcript using small interfering RNA in primary murine and bovine fibroblast cells. Molecular Reproduction and Development, 2005, 72: 311-319.
26 H.J. Unwalla, M.J. Li, J.D. Kim, et al., Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nature Biotechnology, 2004, 22: 1573-1578.
27 D. Grimm, K. Pandey, M.A. Kay, Adeno-associated virus vectors for short hairpin RNA expression. Methods in Enzymology, 2005, 392: 381-405.
28 P. Preecha, Y. Papin, and S. Senee, Drug targeting model of composite gold-tourmaline for cells enhancing applications. Nano Biomed. Eng, 2015, 7(2): 38-46.
29 W. Zhang, H. Yang, X. Kong, et al., Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nature Medicine, 2005, 11(1): 56-62.
30 V. Schramke, R. Allshire, Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science, 2003, 301: 1069-1074.
31 V. Indu, P. Sebastian, F. Taylor, et al., Quantum dot conjugated magnetic nanoparticles for targeted drug delivery and imaging. Nano Biomed. Eng., 2016, 8(1): 24-38.
32 I.J. Majoros, T.P. Thomas, C.B. Mehta, et al., Poly(amidoamine) dendrimer-based multifunctional engineered nanodevice for cancer therapy. J. Med. Chem, 2005, 48: 5892-5899.
33 J. Zhang, F. Xia, Y. Yang, et al., Human CIK cells loaded with gold nanoprisms as theranostic platform for targeted photoacoustic imaging and enhanced immuno-photothermal combined therapy. Nano Biomed. Eng., 2016, 8(3): 112-127.
34 Y.-S. Chen, D.-X. Cui, Non-spherical gold nanoparticles: tumor imaging and therapy, Nano Biomed. Eng., 2013, 5(4), 160-167.
35 T.B. Huff, L. Tong, Y. Zhao, et al., Hyperthemic effects of gold nanorods on tumor cells. Nanomedicine, 2007, 2: 125-132.
36 X.H. Huang, I.H. El-Sayed, W. Qian, et al., Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc, 2006, 128: 2115-2120.
37 B. Pan, F. Gao, L. Ao, et al., Controlled self-assembly of thiol-terminated poly(amidoamine) dendrimer and gold nanoparticles. Colloids and Surfaces A: Physicochem. Eng. Aspect, 2005, 259: 89–94.
38 I.J. Majoros, A. Myc, T.T homas, et al., PAMAM dendrimer-based multifunctional conjugate for cancer therapy: synthesis, characterization, and functionality. Biomacromolecules, 2006, 7: 572-579.
39 C. Sun, C. Nettesheim, Z. Liu, et al., Solution structure of human survivin and its binding interface with smac/diablo. Biochemistry, 2005, 44: 11-17.
40 S. Fuessel, J. Herrmann, S. Ning, et al., Chemosensitization of bladder cancer cells by survivin-directed antisense oligodeoxynucleotides and siRNA. Cancer Lett, 2006, 232: 243–254.
41 L.R. Hirsch, R.J. Stafford, J.A. Bankson, et al., Nanoshell-mediated near-infrared thermal therapy of tumors under magnetic resonance guidance. Proc. Natl.Acad. Sci. U.S.A, 2003, 100: 13549-13554.
42 H. Takahashi, T. Niidome, A. Naria, et al., Gold nanorod-sensitized cell death: microscopic observation of single living cells irriadiated by pulsed near-infrared laser light in the presence of gold nanorods. J. Am. Chem. Soc, 2006, 128: 2115-2120.
43 N.L. Rosi, D.A. Giljohann, C.S. Thaxton, et al., Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science, 2006, 312: 1027-1030.
Copyright© 2017 Chenlu Li, Fangfang Xia, Kan Wang, Can Wang, Ping Xu, Hong Zhang, Jinping Wang, Asahi Toru, Jian Ni, and Daxiang Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.