Research Article
Susceptibility and Antimicrobial Resistance of Genital Ureaplasma Parvum
Ghofran Kadhim Al-khafaji *
Technical Institute of Samawh, Al-Furat Al-Awsat University, Al-Samawah, Iraq.
* Corresponding author. E-mail: ghofran.alkhafaji1@gmail.com
Received: Aug. 1, 2017; Accepted: Sep. 7, 2017; Published: Sep. 27, 2017
Citation: Ghofran K. Al-khafaji, Susceptibility and Antimicrobial Resistance of Genital Ureaplasma Parvum. Nano Biomed. Eng., 2017, 9(3): 236-241.
DOI: 10.5101/nbe.v9i3.p236-241.
Abstract
The object of this study concentrated on investigating the antimicrobial susceptibilities of Ureaplasma parvum isolates to determine the most suitable antibiotic for treating the infection. In total, 35 samples of Ureaplasma parvum isolates were included in this study. Antibiotic susceptibility was studied by broth dilution method which was for the purpose of susceptibility testing of serovar isolates of Ureaplasma parvum against eight antibiotics. The results revealed the serovar 3 isolates were fully resistant (100%) to gentamicin, azithromycin and erythromycin while susceptible at the rates of 80% to doxycycline, 60% to levofloxacin and 60% to clarithromycin. Serovar 14 isolate was revealed fully susceptible (100%) to clarithromycin, ciprofloxacin and doxycycline, while fully resistant (100%) to gentamicin and azithromycin. Serovar 1 and serovar 6 were showed to be fully resistant (100%) to azithromycin and gentamicin. Sevorar 1 was susceptible to at the rates of 70% to doxycycline, 60% to tetracycline, 90% to ciprofloxacin, 70% to levofloxacin, 70% to erythromycin and 70% to clarithromycin. Serovar 6 was susceptible at the rates of 80% to doxycycline, 100% to tetracycline, 100% to ciprofloxacin, 80% to levofloxacin, 80% to erythromycin and 80% to clarithromycin. These results evidently demonstrated that doxycycline, clarithromycin and levofloxacin should be the preferred drug when empirical treatment was required.
Keywords: Serovar; Susceptibility; Resistance; Suitable antibiotics; Isolates; Ureaplasma parvum; Empirical treatment; Antimicrobial; Broth dilution method; Mutation
Introduction
The administration of antimicrobial agents to pregnant women with preterm rupture of the membranes (PROM) may extend the gestation period and decrease the risk of associated complications and neonatal infections. The antimicrobial agent of choice should be considered carefully, as some agents are teratogens - i.e. the agent can cause malformation or functional damage to an embryo or fetus or may have toxic effects on the neonate [1]. Macrolides are often used empirically because of tetracyclines (TETs) and fluoroquinolones being contraindicated in pregnancy. However, the amniotic sac is not effectively penetrated by erythromycin (ERY) and Ureaplasma parvum (U. parvum) are not eradicated from the vagina or cervix by this agent. Newer macrolides (e.g. azithromycin (AZM) and clarithromycin (CLR)) allow better tolerability and the once daily dosing benefit can increase compliance. Treatment with AZM was equally successful, compared to ERY but with fewer side effects [2]. Ureaplasma species (Ureaplasma spp.) contains one or more integrase recombinase genes. Some serovars contain transposases or its remnants and some phage related proteins. The tetM gene was identified as part of a Tn916 transposon, in serovar 9 (SV9) which has acquired TET resistance. A report covering the years 2000 - 2004 from several states in the USA showed that 45% of unique clinical isolates of Ureaplasma spp. contained tetM and were TET-resistant. [3]. The possibility of exchange of DNA between U. parvum and other pathogen within urogenital tract may contribute to gene transfer, which may promote antibiotic resistance in such pathogens [4]. There is only limited information on resistance development of U. parvum. U. parvum is innately resistant to antibiotics which act on cell wall components (the beta lactams). Ureaplasma spp. have natural resistance to lincosamides (e.g. clindamycin) [5]. Other observed resistance to macrolides is associated with mutations in the 23S rRNA gene, while resistance to TET is associated with the presence of the moveable tet M transposon. The tet M gene encodes a protein which binds to ribosomes, and in the case of Ureaplasma spp., it has been demonstrated to be associated, on the chromosome, with Tn916, a conjugative transposon [6]. Previous studies suggested that Ureaplasma spp. resistance to quinolone was mainly due to the mutants of target enzyme-DNA helicase; the residues 68-107 areas were the quinolone-resistant areas (quinolones regions of drug-resistance, QRDR) [7]. Besides beta lactams, U. parvum also shows resistance to sulphonamides, trimethoprim and rifampicin. Resistance to rifampicin is attributed to the presence of a single amino acid at position 526 of the beta sub unit of RNA polymerase. In a recent study conducted by Dhawan et al. [5] involving patients with infertility and genital discharge, all isolates of U. parvum isolates were susceptible to DOX and josamycin; 77% of the isolates were susceptible to ofloxacin and 71% to AZM. Though most studies reported lower resistance rates for TET (< 5%), a recent study by Redelinghuys et al. [2] demonstrated only 27% susceptibility of U. parvum isolates to TET. In a study by Zhu et al. [8] in Shanghai, biovar 1 (Ureaplasma urealyticum) showed high sensitivity rates (above 90%) to all antimicrobial agents; but biovar 2 (U. parvum) maintained higher sensitivities (above 95%) only to DOX and minocycline. In fact, only a small number of biovar 2 strains were sensitive to roxithromycin and quinolones. And U. parvum was shown to be resistant to TET and DOX, because the streptococcal tetM gene transposon was carrying TET-resistant genes (tetM). [20]. All strains of U. parvum were susceptible to josamycin and miocamycin and resistant to trimethoprim, sulfonamides and rifampicin because they did not synthesize folic acid. [20]. U. parvum had higher mutation rate than conventional bacteria which meant it could rapidly develop resistance to drugs including quinolone, ERY and AZM. Resistance developed in U. parvum by point mutation in parC (Pro-57 Leu), and two novel mutations in parE (Ile-73Thr and a methionine insertion at codon 86) were found in an ofloxacin-resistant strain [9, 10, 15].
Experimental
Antimicrobial agents
Antimicrobial susceptibility testing was performed against eight antimicrobial agents that included fluoroquinolones, macrolides, TETs and aminoglycosides, as shown in Table 1 [2, 10]. Each antimicrobial agent was prepared according to the manufacturers’ instructions at a stock concentration of 250 mg / 5 mL.
Table 1 Antibiotics used in antibiotic susceptibility testing.
|
Antibiotics |
Concentration (mg) |
Company /origin |
|
Ciprofloxacin |
250 |
Ajanta / India |
|
Levofloxacin |
250 |
Ajanta / India |
|
Azithromycin |
250 |
Kontam /China |
|
Erythromycin |
250 |
Ajanta / India |
|
Clarithromycin |
250 |
Ajanta / India |
|
Doxycycline |
100 |
Ajanta / India |
|
Tetracycline |
250 |
Ajanta / India |
|
Gentamicin |
120 |
Menarini /Italy |
Antibiotic susceptibility testing
Antibiotics susceptibility was determined by the standard broth dilution method by using Ibtisam habeeb (IH) broth medium containing serial dilution of antibiotics and antibiotics. The control broths were inoculated with bacterial suspension inoculation in IH broth medium.[6]. The inoculated tube was incubated 2 days aerobically at 37 ℃, then spread on IH agar medium and incubated anaerobically at 37 ℃, and read after 24 h under compound light microscope (Olympus Japan) at 10× magnification. The Minimum inhibitory concentration (MIC) was defined as the lowest antibiotics concentration which inhibited the visible growth of U. parvum on the agar plate; the tentative breakpoints used were susceptible (S) and resistant (R).
Procedure of antimicrobial susceptibility testing by broth dilution method
A fixed amount of bacteria was cultured in IH broth medium in sterile test tubes. The concerntrations were numbered for the addition of antibiotic. [9]. Antibiotics were added to make the concentrations in an ascending order, so that the first tube (No.1) contained a zero concentration of antibiotics (control), followed by the second tube (No. 2) that had lower concentration of the antibiotic, and followed by the third tube (No. 3) which contained double the concentration in the tube No. 2. [10]. All the tubes were incubated at the optimum temperature of 37 ℃, then the incubation was investigated, and the growth of bacteria was identified in terms of turbidity. The clear tubes indicated lack of growth of the bacteria as a result of the effectiveness of antibiotics. As in this study U. parvum during its growth did not appear any turbidity in broth, it was kept in the IH agar medium and incubated anaerobically at 37 ℃ for 24 h. Then the plates were investigated to identify the growth of bacteria in terms of colonies. The clear plates indicated lack of growth of the bacteria as a result of the effectiveness of antibiotics. Calculation: To prepare the concentration gradient of the antibiotics 0, 4, 8, 16, 32, 64, 128, 256… μg/mL, 250 mg of ciprofloxacin (CIP) was dissolved in 5 mL distilled water (DW) thus making a concentration of 250 mg / 5 mL, i.e. 50 mg / 1 mL. The units were turned into microgram: 50 × 1,000 = 50,000 μg/mL = Stock 1. Stock 2 = 200 μg / 100 mL was prepared from stock 1.
C1V1 = C2V2 50,000 × V1 = 200 × 100 V1 = 0.4
0.4 mL from Stock 1 + 99.6 D.W became Stock 2 = 200 μg / 100 mL. All the concentrations were prepared as required and with the final volume of 2 mL.
0 = C1V1= C2V2 = 0 Controls
4 = 200×V1= 4×2 V1= 0.04 mL
8 = 200×V1= 8×2 V1= 0.08 mL
16 = 200×V1= 16×2 V1 = 0.16 mL
32 = 200×V1= 32×2 V1 = 0.32 mL
64 = 200×V1= 64×2 V1 = 0.64 mL
128 = 200×V1= 128×2 V1 = 1.28 mL
256 = 200×V1= 256×2 V1 = 2.56 mL
Results and Discussion
Many reports have suggested that U. parvum may be associated with urogenital infections, infertility and adverse pregnancy outcomes [11]. According to previous studies, eight antibiotic agents including DOX, AZM, gentamicin (GEN), CIP, TET, levofloxacin (LVX), ERY and CLR were tested on 35 samples of U. parvum, as they are the major antibiotics used in the treatment of genital tract infection caused by U. parvum. A further purpose of choosing these antimicrobial agents was they are conventionally being used for the routine treatment of sexually transmitted infections [6, 9, 10, 12]. In addition, a new macrolide CLR was also tested. The results are revealed in Table 2, showing that U. parvum isolates were 80% susceptible to DOX (MIC: 8 μg/mL), 71.4% to CLR (MIC: 8 μg/mL), 60% to TET (MIC: 8 μg/mL), 42.8% to ERY (MIC: 16 μg/mL), 65.7% to LVX (MIC: 4 μg/mL) and 74.2% to CIP (MIC: 256 μg/mL). Also, the results showed that U. parvum isolates were 91% susceptible to DOX (16 μg/mL), 85.7% to CLR (16 μg/mL), 65.7% to TET (16 μg/mL), 54.2% to ERY (32 μg/mL) and 82.8% to LVX (8 μg/mL). The U. parvum isolates were highly resistant (100%) to CIP (4 - 128 μg/mL), AZM (4 - 256 μg/mL) and GEN (4 - 256 μg/mL) as recorded in this investigation (Fig. 1). The results of the present study were similar to those of other studies on the rates of susceptibility of U. parvum to antibiotics. [5, 9, 12]. U. parvum has been considered susceptible to macrolides; however, in the present study, U. parvum was resistant to AZM and ERY. Similar to the findings of Kilic et al. [7], AZM’s resistance to strains of U. parvum was reported with increasing frequency. The resistance against AZM developed in U. parvum by a point mutation in parC, and two novel mutations in parE were found in an ofloxacin-resistant strain [11,13]. That the strains of U. parvum were resistant to ERY [12] may be because of the mutations in the L22 ribosomal protein which were seen in three strains that were resistant to ERY [4]. No resistance was seen against the new macrolide CLR in U. parvum, and the results were similar to those reported earlier [10, 14]. TET and DOX were the most active agents against U. parvum. This finding was consistent with those of other studies conducted in China [4, 15] and Turkey [7, 16]. The majority of U. parvum serovars were susceptible to DOX, CLR and LVX, while U. parvum serovars were fully resistant (100%) to GEN and AZM [13]. The resistance to GEN and AZM was of intermediate nature in all the U. parvum isolates. The results revealed that SV3 isolates were fully resistant (100%) to GEN, AZM and ERY, while susceptible to DOX in the rates of 80% and 60% to LVX and 60% to CLR. SV14 isolates were revealed to be fully susceptible (100%) to CLR, CIP and DOX, while fully resistant (100%) to GEN and AZM. SV1 and SV6 showed full resistance (100%) to AZM and GEN. SV1 was susceptible in the rates of 70% to DOX, 60% to TET, 90% to CIP, 70% to LVX, 70% to ERY and 70% to CLR, while SV6 was susceptible at the rates of 80% to DOX, 100% to TET, 100% to CIP, 80% to LVX, 80% to ERY and 80% to CLR. The results were shown in Fig. 2. The study further observed that U. parvum SV3 was the most frequent serovar detected in patients and was susceptible to DOX, LVX and CLR [14] while resistant to ERY, AZM and GEN. This finding was consistent with those of other studies [6, 7, 17]. The fluoroquinolones are considered useful in the treatment of Ureaplasma infections as they are potentially effective against pathogenic species and contain strains that are resistant to other drugs such as DOX. Fluoroquinolones are used for treating urogenital infections; they interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV [9]. The results in the present study showed that U. parvum isolate was susceptible to levofloxacin. However, U. parvum was resistant to CIP (fluoroquinolones). Similar rates of resistance to quinolones were observed in clinical isolates of U. parvum in most of the studies [7, 16, 18]. There were some reports of fluoroquinolone-resistant U. parvum in the field of urology in Japan [11]. Resistance against fluoroquinolones in U. parvum may occur to some degree due to the widespread use of these drugs for the treatment of respiratory and urogenital infections. U. parvum resistance to fluoroquinolones has been attributed to substitution mutations, principally in gyrA and parC genes, and to a lesser extent in gyrB and parE genes of the DNA gyrase / topoisomerase IV complex [17]. Six mutations in parC gene, five mutations in parE and one mutation in gyrA which resulted in amino acid substitutions were identified in U. parvum. There were only three silent mutations observed in gyrB. The gyrA mutation that resulted in the substitution of glutamine for lysine at amino acid 103 in U. parvum may be near enough to the tyrosine active site of the protein at amino acid 122 to contribute to the resistance against CIP as observed [6, 7]. However, a high rate of resistance to GEN was observed for U. parvum. GEN belongs to the group of aminoglycosides which are not active against anaerobes bacteria. Three mechanisms of resistance have been recognized, namely ribosome alteration, decreased permeability and inactivation of the drugs by aminoglycoside modifying enzymes. The latter mechanism is of most clinical importance since the genes encoding aminoglycoside modifying enzymes can be disseminated by plasmids or transposons [2, 6, 19]. Ribosome alteration can result in single mutations in chromosomal genes encoding ribosomal proteins: rpsL (or strA), rpsD (or ramA or sud) and rpsE (eps or spc or spcA) [15]. Decreased permeability alteration in the aminoglycoside transport system, inadequate membrane potential and modification in the LPS (lipopolysacchaccarides) phenotype can result in a cross resistance to all aminoglycosides. In the activation of aminoglycosides, these enzymes are classified into three major classes according to the type modification: acetyltransferases (AAC), nucleotidyltransferases or adenyltransferases (ANT ) and phosphotransferases (APH) [7].
Table 2 Antibiotics used in antibiotic susceptibility testing
|
Antibiotics / total number (35) |
MIC (μg/mL) |
Rate susceptibility |
|
Ciprofloxacin |
256 |
26 (74.2%) |
|
Levofloxacin |
4 |
23 (65.7%) |
|
Azithromycin |
- |
0 |
|
Erythromycin |
16 |
15 (42.8%) |
|
Clarithromycin |
8 |
25 (71.4%) |
|
Doxycycline |
8 |
28 (80%) |
|
Tetracycline |
8 |
21 (60%) |
|
Gentamicin |
- |
0 |
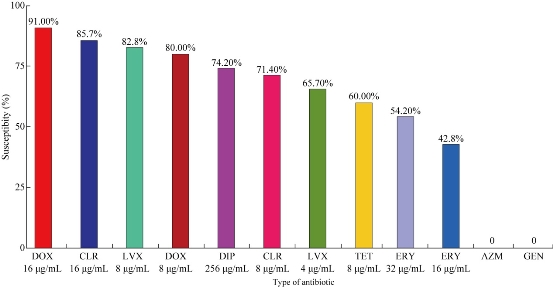
Fig. 1 Susceptibility results of U. parvum to different antibiotics.
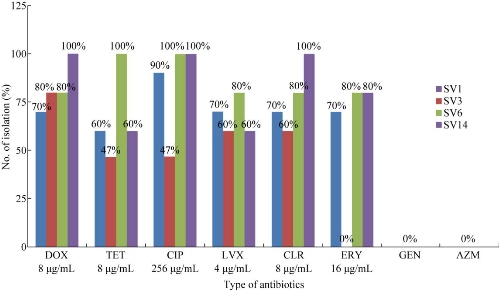
Fig. 2 Susceptibility (MIC) of serovars to different antibiotics.
Conclusions
These results evidently indicated that DOX, CLR and LVX should be the drug of first choice when empirical treatment is required. The results revealed in the present study indicated that U. parvum colonizing the genital tract epithelium might come form biofilms that protected organisms from host defenses and antibiotic treatment. To confirm the biologic relevance of the current in-vitro studies, biofilm-formation of clinical U. parvum isolates should be tested with in-vivo experimental models. Treatment of U. parvum infection is imperative to prevent the occurrence of complications. Empirical therapy is important in the treatment of U. parvum infections.
Conflict of Interests
The authors declare that no competing interest exists.
Copyright© 2017 Ghofran Kadhim Al-khafaji. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.