Research Article
High-purified Isolation and Proteomic Analysis of Urinary Exosomes from Healthy Persons
Meng Yang 1, Xiao Zhi 1, Yanlei Liu 1, 2, Tianliang Li 1, Gabriel Alfranca 1, Fangfang Xia 1, Chenlu Li 3, Jie Song 1*, Daxiang Cui 1, 2
1 Institute of Nano Biomedicine and Engineering, Shanghai Engineering Research Center for Intelligent Instrument for Diagnosis and Therapy, Key Laboratory for Thin Film and Microfabrication of Ministry of Education, Department of Instrument Science and Engineering, School of Electronic Information and Electronical Engineering, 800 Dongchuan Road, Shanghai Jiao Tong University, Shanghai 200240, P. R. China.
2 National Center for Translational Medicine, Collaborative Innovational Center for System Biology, Shanghai Jiao Tong University, 800 Dongchuan RD, Shanghai 200240, P. R. China.
3 Key Laboratory of Laboratory Medicine, Ministry of Education of China, Zhejiang Provincial Key Laboratory of Medical Genetics, School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, Zhejiang 325035, P. R. China.
* Corresponding authors. E-mail: dxcui@sjtu.edu.cn; sjie@sjtu.edu.cn
Received: Sep. 10, 2017; Accepted: Sep. 20, 2017; Published: Sep. 27, 2017
Citation: Meng Yang, Xiao Zhi, Yanlei Liu, Tianliang Li, Gabriel Alfranca, Fangfang Xia, Chenlu Li, Jie Song, and Daxiang Cui, High-purified Isolation and Proteomic Analysis of Urinary Exosomes from Healthy Persons. Nano Biomed. Eng., 2017, 9(3): 221-227.
DOI: 10.5101/nbe.v9i3.p221-227.
Abstract
Urinary exosomes containing specific biomarkers have recently been considered as novel potential non-invasive candidates for renal disease diagnosis. However, the development of urinary exosomes in basic research and their subsequent diagnostic application are impeded by the lack of an efficient isolation method. One of the main challenges during urinary exosomes isolation is how to remove a large number of Tamm Horsfall proteins (around 92 kDa) and other biological components from exosome enrichment mixture. Herein, we report a facile and low-cost isolation method for highly-purified human urinary exosomes based on dialysis. The key protocol for exosome isolation includes only two steps: (1) Healthy person urines were collected. 10 mL urine in 300 kDa dialysis tubes was firstly dialyzed in phosphate-buffered saline solution three times for sequential nine hours; (2) The dialysis suspension was concentrated to 200 μL by using 100 kDa ultracentrifuge tubes to achieve urinary exosome isolation. For verification, the concentrated solution was examined by western blot, transmission electronic microscopy, atomic force microscopy and qNano, which demonstrated the highly-purified urinary exosomes were present. Furthermore, a total of 359 proteins were identified by the proteomic analysis of purified urinary exosomes from healthy persons. Those results demonstrated that highly-purified urinary exosomes could be achieved by our isolation method; 359 proteins were identified from healthy persons. Further works will focus on screening and identifying disease-related biomarkers from human urine exosomes for clinical diagnosis.
Keywords: Urinary exosomes; Hamm-horsfall protein; Dialysis; Proteomic analysis
Introduction
In recent years, liquid biopsy has made great advance. Exosomes from different organs or cells exhibit great potential in applications such as disease diagnosis and therapy. Urinary exosomes have been found to own great potential application in disease diagnosis, therapy and disease molecular mechanism. Urinary exosomes are 30 - 150 nm membrane vesicles released by all cell types along the structure of the kidney, which was firstly described by Pisitkun et al. in 2004 [1]. In the last decade, urinary exosomes have received a considerable attention for their potential as non-invasive renal disease biomarkers due to their enrichment protein [2], mRNA [3] and microRNA complex [4]. However, researches on urinary exosomes have been hampered by lacking a facile and low-cost isolation technique for highly-purified exosomes [5]. Currently, the existing methods are mainly based on ultracentrifugation [1], ultrafiltration [6], size exclusion [7], polyethylene glycol (PEG) [8] or commercial precipitation reagent [9]. The advantages and disadvantages of the different methods have been compared in previous reports [10, 11]. In a word, those methods based on size exclusion or ultracentrifugation cannot achieve highly-purified exosomes, while the methods based on precipitations provide even less purified exosomes [11]. In urine, Tamm-Horsfall proteins (THP, around 92 kDa) are the main type of mixtures, which are essential for the isolation of urinary exosomes [12]. Musante et al. reported the use of 1000 kDa dialysis tubes to exclude THP by hydrostatic pressure and then exosomes were recovered by 200,000 g ultracentrifugation [13]. However, the promotion of this method which required the ultracentrifugation step is limited in ordinary labs due to the high-cost centrifuge equipment. More importantly, the tough process during ultracentrifugation may also induce exosomes to aggregate [14] or crack [15]. To overcome the above shortcomings, we developed a facile and low cost method for highly-purified urinary exosomes isolation by using 300 kDa dialysis tubes and without ultracentrifugation. Schematic 1 illustrates the protocol of urinary exosome isolation based on dialysis - a common and easy performed lab technique. Then, we analyzed proteomic profiling of urinary exosomes from healthy persons, which lay foundation for further identifying biomarkers associated with kidney diseases by comparing it with proteomic profiling of urinary exosomes from kidney disease patients.
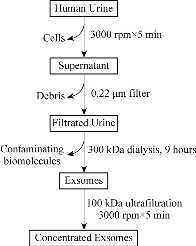
Schematic 1 The protocol of urinary exosomes isolation.
Experimental
Urine collection
50 mL of first-void urine was collected from three healthy human volunteers in the morning and processed immediately after collecting without storage.
Isolation of urine exosomes by dialysis
Urine samples were centrifugated at 3,000 rpm for 10 min to remove whole cells and other large debris at 4 C. The supernatants were collected and filtrated by 0.22 μm filtration (Millipore, USA). Then, 10 mL filtrated urine was added to 300 kDa dialysis tubes (Spectrumlabs, USA) and dialyzed in phosphate-buffered saline (PBS) for a total of 9 h with replacing fresh PBS every 3 h. The dialyzed urine was collected and 100 kDa ultrafiltration tubes (Millipore, USA) were used to concentrate the exosomes to the volume of around 200 μL.
Nanodrop analysis concentrations
Nanodrop (Thermo Fisher Scientific, USA) was used to measure proteins and nucleic acid concentrations with PBS as control.
Transmission electron microscopy
A volume of 10 μL exosomes was added on carbon-coated 200-mesh copper grid and removed by filter paper after 1 min. 5 μL uranyl acetate was then applied on the copper grid for 1 min for counterstaining and removed by filter paper. The samples were examined with 120 kV biology transmission electron microscope (Tecnai G2 spirit Biotwin).
Atomic force microscopy
The isolated exosomes were diluted (1:100) in deionized water. The diluted exosomes were added to freshly cleaved mica sheets and dried under a gentle stream of nitrogen. Samples were analyzed by environment control scanning probe microscope in tapping mode.
Quantification of exosomes particles
We quantified the number of urinary exosome particles using qNano device (Izon Science, New Zealand).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis and western blot
SDS-PAGE was used to analyze the dialysis efficiency. Briefly, 15 μL samples were used for SDS-PAGE analysis and the dispersed proteins were detected by coomassie brilliant blue staining. TSG101 (Abcam, UK) and CD9 (Abcam,UK) antibodies were used in the western blot analysis.
Gel-based proteomic analysis
Entire gel lanes were cut into 1.5 mm bands, proteins in-gel were digested by trypsin and extracted, then analyze by Nano-Liquid Chromatography (Thermo Fisher Scientific, USA).
Results and Discussion
Removing contaminating biomolecules
To achieve highly-purified exosomes, it is essential to remove outside contaminating biomolecules from exosomes. As shown in Fig. 1(a)-(c), the concentrations of proteins, DNA and RNA in fresh urine were 64 mg/mL, 2,995 ng/μL and 2,353 ng/μL respectively. After 3 h dialysis the concentrations of these biomolecules were reduced significantly and decreased to 0.05 mg/mL, 3.4 ng/μL and 1.03 ng/μL by the end of 9 h dialysis respectively. Another indicator of those phenomena was the color changing of specimens. After 3 and 9 h dialysis, the urinary color changed from clear yellow to weaker yellow and finally colorless (Fig. 1(d)). Those results demonstrated that dialysis could remove contaminating biomolecules efficiently.
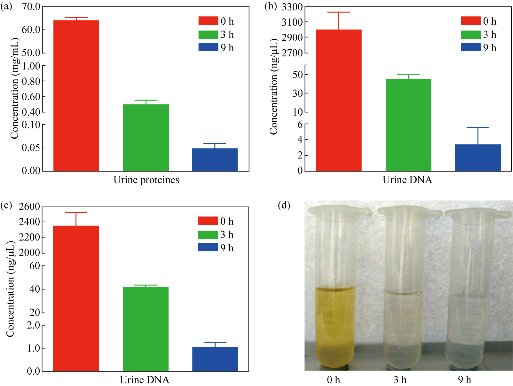
Fig. 1 The concentrations of urinary biomolecules and the urinary color at different dialysis times. (a) Proteins; (b) DNA; (c) RNA concentrations; and (d) Color of fresh urine and urine after dialysis for 3 and 9 h.
Protein reduction and western blot analysis of urinary exosomes
To demonstrate the reduction of proteins, SDS-PAGE gels were stained with coomassie brilliant blue. As shown in Fig. 2(a), after 3 h dialysis, the protein presences was dramatically reduced, especially for THP. As both CD9 and TSG101 were generally used as biomarkers of urinary exosomes [12]. CD9 and TSG101 expression could be detected in the final solution, which indicated exosomes were present in the solution (Fig. 2(b)).
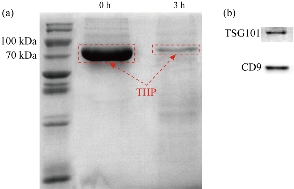
Fig. 2 Coomassie brilliant blue staining SDS-PAGE of urinary proteins and urinary exosomes western blot. (a) Proteins reduced especially THP in red dotted box after dialysis 3 hours; (b) Immunoblots of urinary exosomes against TSG101 and CD9 proteins
Transmission electronic microscopy (TEM) of urinary exosomes
Urine after 3 h dialysis was prepared for TEM imaging. The TEM results demonstrated that polymeric filament formed by THF was retained in urine [12] (Fig. S1). However, after 9 h dialysis, purified exosomes with typical “cup” structure were achieved without the aforementioned filaments as shown in Fig. 3. The results indicated that 9 h was enough to isolate purified urinary exosomes.
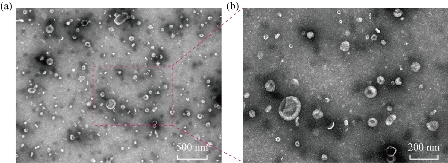
Fig. 3 TEM images of urinary exosomes. The urinary exosomes were identified by (a) size and (b) shape (cup).
Atomic force microscopy (AFM) and qNano of urinary exosomes
The exosomes characterizations were also carried out by AFM and qNano. As shown in Fig. 4(a) and (b), the AFM results showed the structures of urinary exosomes. The qNano measurements demonstrated that the mean diameter of urinary exosomes was around 102 nm as shown in Fig. 4(c).
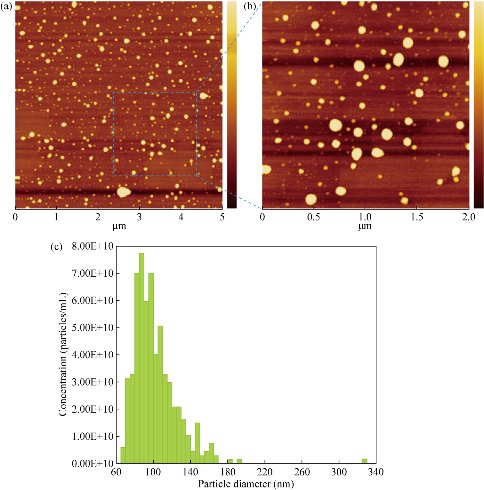
Fig. 4 Characterizations of urinary exosomes by AFM and qNano. AFM images of urinary exosomes with (a) low magnification and (b) high magnification, as well as (c) qNano of urinary exosomes.
Urinary exosomes proteomics analysis
The urinary exosomes were isolated from healthy persons urine. The urinary exosomes proteomics analysis was carried out and a total of 359 proteins were identified as shown in Fig. 5 and Table S1. These proteins mainly included 17 keratin, 15 growth factors, 25 Ig protein components, 68 enzymes, 40 adhesive molecules, 31 inhibitors, 12 glycoproteins, 11 lipoproteins and complement C proteins, 12 heat shock proteins, 15 ion channel proteins, 34 member proteins, some signal transduction proteins, etc. Especially, there existed some special antigens such as CD44 antigen, CD59, prostate stem cell antigen and suppressor of cytokine signaling 2. The proteins will facilitate the application of exosomes in renal disease research such as screening biomarkers. This study aimed to provide a facile and low cost isolation method for high purified urinary exosomes. Several exosomes isolation methods have been described in previous reports [7, 8, 15, 16]. The key point of those methods was how to remove the contaminating molecules outside the exosomes, especially the THP proteins [12]. Dialysis was a good choice to achieve the goal. Our results demonstrated that the THP protein content was remarkably removed by dialysis. The obtained exosomes were present in the original exosomes with specific proteins CD9 and TSG101 as indicators. The advantages of dialysis compared with other more aggressive techniques are: high yield of pure exosomes, less damage without ultracentrifugation process, facile process and low cost. AFM has been used to compare the structures of the exosomes isolated by different methods; the ultracentrifugation method has provided individual, regular and fairly rounded structures [17]. Our AFM result showed that the structure of the exosomes obtained was similar to the ultracentrifugation method but without the risk of aggregation [14] and disruption [15] by avoiding high speed ultracentrifugation steps. In this paper, 300 kDa dialysis tubes were used for the 30 nm pore size [18], which guaranteed that most exosomes were retained in the dialysis tube, while other smaller molecules with weights lower than 300 kDa could be quickly removed. The TEM results demonstrated the high yield pure exosomes were achieved by our method. Dialysis is an easy-to-handle technique that requires no special training which improves the operability of our method. In addition, the dialysis tube is less expensive than the commercial exosomes isolation kit, and the dialysis tube can be reused several times which reduces the cost of exosomes isolation. Up to date, exosomes have also been confirmed to be associated with immune regulation [19], to mediate communication within the tumor microenvironment [20] and to be naturally-equipped nanocarriers for drug delivery [21]. MiRNA-221 in exosomes from bone marrow mesenchymal stem cells could activate oncogenic activity in gastric cancer [22]. Although gastric cancer biomarkers and theranostics have made great advance [23, 24, 25, 26]; some miRNAs have also been found to be associated with gastric cancer stages [27], how the exosomal proteins involved in the progression of cancer still lacks report. Li, et al. reported that exosomal proteins were used for cancer diagnosis, were potential biomarkers [28], and could also be used for cardiac angiogenesis [29], tumor therapy [30] and (hepatitis B virus) HBV therapy [31]. Tumor cell-derived exosomes could be used for tumor therapy [32]. In this study, we confirmed that there existed 359 kinds of proteins in urinary exosomes from healthy persons. The proteins belonged to several types and owned different functions, which can be used for screening disease biomarkes. If exosomes combine with nanotechnology, applications of exosomes in diagnosis and therapy will be improved greatly [33]. Our investigation lays foundation for further clarifying the theranostic mechanism of exosomes.
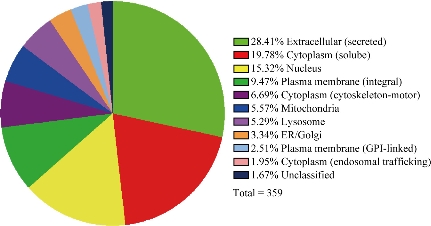
Fig. 5 Pie chart representing the protein categories of urinary exosomes.
Conclusions
In summary, this study provided a facile and low-cost isolation method for high purified exosomes with a potential to facilitate applications of exosomes in disease research.
Acknowledgements
This work was supported by the National Key Basic Research Program (973 Project) (No.2015CB931802), National Natural Scientific Fund (No. 81225010, 81327002 and 31100717), 863 Project of China (2014AA020700) and Shanghai Science and Technology Fund (No. 13NM1401500 and 15DZ2252000).
Conflict of Interests
The authors declare that no competing interest exists.
Supporting Information
Additional information not shown in the main text are provided.
References
Copyright© Meng Yang, Xiao Zhi, Yanlei Liu, Tianliang Li, Gabriel Alfranca, Fangfang Xia, Chenlu Li, Jie Song, and Daxiang Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.