Research Article
Extracellular Biosynthesis of Silver Nanoparticles Using Fungi and Their Antibacterial Activity
Paula Sanguiñedo 1, Raluca María Fratila 2,3, María Belén Estevez 1, Jesús Martínez de la Fuente 2,3, Valeria Grazú 2,3**, Silvana Alborés 1*
1 Área de Microbiología, Departamento de Biociencias, Facultad de Química, Universidad de la República, Montevideo, Uruguay.
2Instituto de Ciencia de Materiales de Aragón, CSIC, Zaragoza, España.
3Centro de Investigación Biomédica en red en Bioingenieria Biomateriales y Nanomedicina (CIBER-BBN), Zaragoza, Spain.
* Corresponding author: Email: salbores@fq.edu.uy; Tel.: + 598 2 9244209; Fax: +598 2 9241906.
** Co-corresponding author: E-mail: vgrazu@unizar.es; Tel: +34 876 555361; Fax: +34 976 762776.
Received: Jan. 12, 2018; Accepted: May 22, 2018; Published: Jun.1, 2018
Citation: Paula Sanguiñedo, Raluca María Fratila, María Belén Estevez, Jesús Martínez de la Fuente, Valeria Grazú, and Silvana Alborés, Extracellular Biosynthesis of Silver Nanoparticles Using Fungi and Their Antibacterial Activity. Nano Biomed. Eng., 2018, 10(2): 165-173.
DOI: 10.5101/nbe.v10i2.p165-173.
Abstract
Silver nanoparticles have particular properties that contribute to their very promising applications, novel in various fields of science, such as the development of biosensors, the diagnosis and treatment of cancer, the controlled release of drugs and the antimicrobial potential. The biological synthesis of nanoparticles is of great interest over other physical and chemical methods because the use of toxic chemicals and drastic reaction conditions are avoided. The extracellular biosynthesis using fungi could also make downstream processing much easier than the intracellular biosynthesis. One of the main applications of silver nanoparticles is their antimicrobial activity. Several studies have demonstrated the bactericidal properties of silver nanoparticles are different from silver ions, and that they are strongly influenced by their shape, size, concentration and colloidal state. In the present work, the ability of fungal strains from Uruguay to synthesize silver nanoparticles was studied. Eight fungi were able to synthesize nanoparticles. An extensive physicochemical characterization of the nanoparticles was carried out including ultraviolet-visible spectroscopy, transmission electron microscopy, dynamic light scattering, zeta-potential and gel electrophoretic mobility. According to the characterization and colloidal stability results, nanoparticles from three fungi were selected for antimicrobial activity assays. All nanoparticles were able to inhibit Escherichia coli growth, demonstrating their potential as effective antibacterial agent for use in biomedical applications.
Keywords: Silver nanoparticles; Fungi; Biosynthesis; Antibacterial activity
Introduction
Silver nanoparticles have attracted considerable attention due to their diverse properties and applications such as controlled drug delivery, biosensors, diagnostic and treatment of cancer, or antimicrobial agents [1-3]. Nanoparticles are clusters of atoms in the size range of 1- 100 nm. Their high surface area to volume ratio give nanoparticles new mechanical, chemical, electrical, optical, magnetic, electro-optical and magneto-optical properties that are different from their bulk properties [2, 4]. In recent years, the green approach of nanoparticle synthesis by biological entities has been gaining interest over various other physical and chemical methods. The biological method of synthesis of nanoparticles has proved to be better than some chemical or physical methods due to slower kinetics, which offers a better control over crystal growth and elimination of hazardous chemicals, making it an eco-friendly method [5]. These methods of biosynthesis include the intracellular or extracellular production of nanoparticles of different elements by a variety of microorganisms and plant extracts [6, 7]. However, the extracellular biosynthesis using fungi could also make downstream processing much easier than the intracellular biosynthesis. In addition, fungi have been known to secrete high amounts of bioactive substances, which make them very suitable for large-scale production. Previous reports showed that these active substances secreted by fungi played important roles such as reducing agents and capping agents in the reaction; however, the detailed mechanism of biosynthesis of silver nanoparticles has not been well elucidated [8].The objective of the present study was to evaluate the ability of fungal strains from Uruguay to synthesize silver nanoparticles and to determine their antimicrobial activity. To our knowledge, synthesis from Punctularia atropurpurascens, Botrytis cinerea and Gymnopilus spectabilis species had not been reported so far, so we encouraged the evaluation of these fungi in order to find new potential species and to contribute to a better understanding of the biosynthesis process. Although there had been previous reports of biosynthesis with some of the other species evaluated, the strains, the conditions of the culture and reaction performed in this work were different from the other reports. To this aim, the production of silver nanoparticles using the extracellular cell-free filtrates of these fungi was investigated. The use of controlled and similar conditions for the nanoparticle synthesis allowed carrying out a comprehensive comparison among them that had not been done so far. In this sense, an extensive physicochemical characterization was carried out including ultraviolet-visible spectroscopy (UV-Vis), transmission electron microscopy (TEM), dynamic light scattering (DLS), zeta-potential(-potential) and gel electrophoretic mobility. Moreover, the antimicrobial activity of silver nanoparticles against Gram-positive and Gram-negative bacteria was evaluated. Another aspect evaluated was their stability against centrifugation and at different pH and ionic strengths. This information was not usually reported despite its importance not only for the downstream processing of the nanoparticles but also for their further biofunctionalization if their intended final use concerns applications where their multifunctionalization is a must.
Experimental
Chemicals and materials
Potato Dextrose Agar,Potato Dextrose Broth and Mueller Hinton Agar were purchased from BD Difco. Silver nitrate and 2-(N-morpholino) ethanesulfonic acid buffer (MES) were from Sigma Aldrich. Agarose was from Lonza. Membrane filters were purchased from Millipore.
Organisms
Strains of Punctularia atropurpurascens (H2126),Botrytis cinerea (14G), Penicillium expansum (14S), Fusarium graminearum (31084), Pleurotus ostreatus (816), Phanerochaete chrysosporium (12G),Alternaria alternata (54027), Rhizopus stolonifer 6227B), Gymnopilus spectabilis (354), Abortiporus biennis (12GC), Ganoderma resinaceum (7187) and Grifola frondosa (14TM) from the Cátedra de Microbiología General Collection (CCMG, Facultad de Química, Montevideo, Uruguay), were used in the present study.
Synthesis of silver nanoparticles
The mycelia were grown in Potato Dextrose Agar at 28 ºC and two plugs of 0.9 cm in diameter were then transferred to 500 mL flasks containing 100 mL Potato Dextrose Broth. Fermentations were carried out at 28°C, with agitation on an orbital shaker operating at 150 rpm for 5 days. The biomass from cultures was harvested by filtration and then washed extensively with sterilized distilled water to remove any remaining media components. Then, wet fungal mycelia were suspended in sterilized distilled water (0.1g/mL) and incubated at 28°C with agitation on an orbital shaker operating at 150 rpm for 72 h. Then, the cell-free filtrate was collected by filtration of this suspension through membrane filter with 0.45 µm pore size. Finally, 50 mL of the cell-free filtrate was added to 50 mL of a 1 mM silver nitrate solution. The mixture was incubated at room temperature in dark. The absorbance spectrum was measured in the range of 250 - 800 nm and the maximum peak was determined, at different times. The reaction was stopped when there was no increase in the maximum absorption peak of silver nanoparticles. The remaining cell-free filtrate was used as control.
Purification of silver nanoparticles
To obtain purified nanoparticles, the samples were centrifuged at 10000 rpm for 10 minutes after reaction. The supernatant was removed and nanoparticles were washed twice using 10 mM 2-(N-morpholino) ethanesulfonic acid buffer (MES) pH 6, by centrifuging the nanoparticles 5 min at 10000 rpm. Then, absorbance peak of purified silver nanoparticles was measured.
Characterization of synthesized silver nanoparticles
UV-Vis
The color changes of reaction mixtures were used as evidence for silver nanoparticles formation. Therefore, 1 mL samples were withdrawn from flasks containing colloidal silver nanoparticles at predetermined time intervals, and the absorbance spectrum was measured in the range of 250 -800 nm.
TEM
For TEM examinations, a single drop (10 µL) of the aqueous solution (0.1 mg/mL) of different nanoparticles was placed onto a copper grid coated with a carbon film (Electron Microscopy Sciences). The grid was left to dry in air for several hours at room temperature. TEM analysis was carried out in a TECNAI T20 electron microscope (FEI) working at 200 kV. The particle average size was evaluated from several micrographs using an automatic image analyzer (Image J Software).
Electrophoretic analysis
A 1% (w/v) agarose gel was prepared by dissolving agarose powder in 0.5X tris–borate–EDTA buffer (TBE). The samples were loaded by mixing 80 µL of nanoparticles in water with 20 µL of glycerol: TBE (1:4). Gels were run for 30 min at 80 V. Visible light was used to detect nanoparticles.
DLS and -potential
A Malvern Instruments Zetasizer ZS-NANO was used for the determination of the hydrodynamic diameter (by DLS) and for the measurement of -potential of the nanoparticles. Samples were prepared at pH 6, in Milli-Q water.For DLS determination, each sample was measured at 25 °C, 10 times, combining 5 runs per measurement. In the case of ζ-potential, each sample was measured at25 °C, 3 times, combining 10 runs per measurement. Results were treated using the Malvern software Zetasizer Nano 7.03.
Colloidal stability assays
The nanoparticles colloidal stability was studied at different pH (5 and 7) and ionic strength (10, 50 and 100 mM buffer MES) conditions by the measurement of the absorbance spectrum.
Antimicrobial assays
Antimicrobial activity was determined according to Clinical and Laboratory Standards Institute (CLSI, 2012). Test organisms used were Staphylococcus aureus (ATCC 6538) and Escherichia coli (ATCC 11105). For Agar Diffusion Method, 20 μL of nanoparticles were applied on 5 mm filter paper discs; Gentamycin (100 μg/mL) and extracellular extract were used as positive and negative control, respectively.
Results and Discussion
Biosynthesis of silver nanoparticles.
From twelve fungal strains evaluated, eight (P. atropurpurascens, B. cinerea, P.expansum, P. ostreatus, P. chrysosporium, R. stolonifer, G. spectabilis, and G. frondosa) were found to reduce the silver salt to nanoparticles. This was observed through a color change in the reaction mixtures and the appearance of absorption bands between 400 and 450 nm, which were indicative of pronounced silver plasmon resonances (Fig. 1). This signalized the formation of silver nanoparticles. Besides, the electronic transitions involving the Ag+ ion gave rise to absorption bands located between 200 and 230 nm, whereas the electronic transitions of metallic Ag0 appeared in the 250-330 nm spectral range [9]. No color change was observed in the cell free supernatant alone used as control. The time course of the biosynthesis of silver nanoparticles (measured at 440 nm) for the eight strains is shown in Fig. 2. This is the first report of synthesis of silver nanoparticles from P. atropurpurascens, B. cinerea and G. spectabilis species. Although there had been previous reports of biosynthesis with the other species evaluated, the strains and the conditions of the culture and reaction were different, so differences in the characteristics of the obtained silver nanoparticles were expected [10]. For example, synthesis of nanoparticles from Alternaria alternata and Fusarium graminearum cultures, grown in chemically-defined media, was previously reported [11, 12]; however, the production of nanoparticles from the culture of these two fungi in nutrient medium was not detected in this work. Furthermore, biosynthesis of silver nanoparticles from biomass of Pleurotus ostreatus grown in 5 g/L malt extract and 10 g/L glucose broth or Penicillium expansum grown in yeast mold (YM) broth, was reported [13, 14]. In our work, both fungi were grown in Potato Dextrose Broth. This difference could explain the different absorption peaks of silver nanoparticles produced in our experiments. As P. expansum nanoparticles showed a high and narrow peak (Fig. 1), P. ostreatus nanoparticles showed a broad and lower absorption peak. As shown in Fig. 2, P. ostreatus showed the lowest absorbance values during the 144 hours evaluated; for this reason, this fungus was not selected for further characterization. Previously, Phanerochaete chrysosporium and Grifola frondosa were evaluated for the production of silver nanoparticles [15, 16]. The reaction was done with the mycelium but not with the extracellular cell-free filtrates as described in the present manuscript.
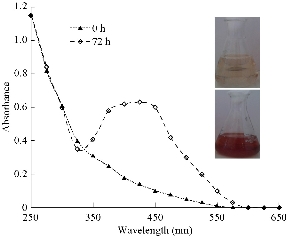
Fig. 1 Color change and UV-Vis absorption spectra obtained for silver nanoparticles synthesized by Penicillium expansum after 72 h of reaction.
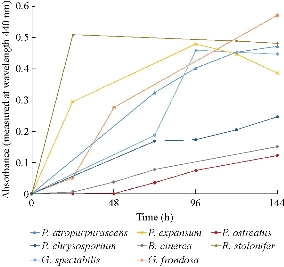
Fig. 2 Time course of biosynthesis of silver nanoparticles with the eight extracts of strains able to produce nanoparticles.
Purification of silver nanoparticles.
All the samples showed strong absorption peaks in the visible region of their absorption spectra at 144 h of reaction except when the mycelia of P. ostreatus was used; thus, the nanoparticles (NPs) obtained using this strain (PoNPs)were discarded for further studies. The rest of the samples were centrifuged and the nanoparticles in the pellet were washed in order to obtain isolated nanoparticles. UV-Vis spectroscopy was used to study the colloidal stability of the purified nanoparticles. This is a straightforward and simple method to assess the stability of metallic nanoparticles, which can be applied to monitor their quality over time as well as evaluating the integrity of the colloidal solution when performing purification steps or surface modifications. If the formation of aggregation states happens, the surface plasmon resonance (SPR) of individual particles becomes coupled, which is accompanied by a red-shift in the spectrum, as well as broadening of adsorption peaks, and a decrease in peak intensities. This information is not usually reported. For example, biosynthesis of silver nanoparticles from R. stolonifer was previously reported, but they were not separated from the reaction medium [17]. After re-supension of the centrifuged nanoparticles, only in the case of the nanoparticles obtained from P. atropurpurascens (PaNPs), P. expansum (PeNPs), P. chrysosporium (PchNPs) and G. spectabilis (GsNPs) the UV-Vis spectra were similar to the ones before centrifuging. A decrease in the peak intensity could be expected due to losses of nanoparticles during their purification. However, neither a red-shift in the spectrum (SPR over 400 nm) nor a significant broadening of adsorption peaks was observed (Fig. 3). This was not the case for nanoparticles obtained from B. cinerea, R. stolonifer and G. frondosa (BcNPs, RsNPs, and GfNPs, respectively), as their UV-Vis spectra showed that the original extinction peak not only decreased in intensity (due to the depletion of nanoparticles) but also broadened (due to the formation of aggregates). Although mechanism of biosynthesis and capping agent are still unknown, these differences in stability of the different biosynthesized nanoparticles (from different fungal strains) could be attributed to the organic shells attached to the inorganic core in a less or more stable way. Unlike some biosynthesized silver nanoparticles reported until now, it was possible with four strains (two of them never used before) to obtain nanoparticles that could resist centrifugation for their separation of the reaction media.
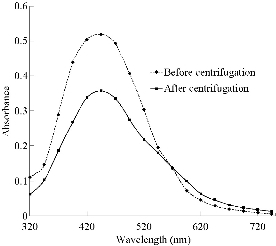
Fig. 3 Use of UV-Vis to assess the colloidal stability of the nanoparticles after a centrifugation step. UV-Vis absorption spectra of PeNps before and after the centrifugation step are shown as example of nanoparticles that resist this purification treatment. As the samples were concentrated due to the centrifugation, the purified nanoparticles were diluted accordingly (5 times) to be compared with the spectra of the non-purified nanoparticles.
Electron microscopy characterization of the nanoparticles
TEM characterization provided further insight into the morphology and particle size distribution profile of the seven different nanoparticles that were precipitated by centrifugation (Fig. 4). The analysis of micrographs using Image J software revealed that the silver nanoparticles were primarily spherical in shape with sizes ranging between 10 and 40 nm. Average size (AS) obtained from each micrograph showed their sizes were between 11 nm (the smallest nanoparticles) and 40 (the biggest nanoparticles).The presence of aggregates in the TEM images could not be related to their actual dispersion state of the nanoparticles in suspension as the drying process of the sample on the TEM grid often introduces artifacts.
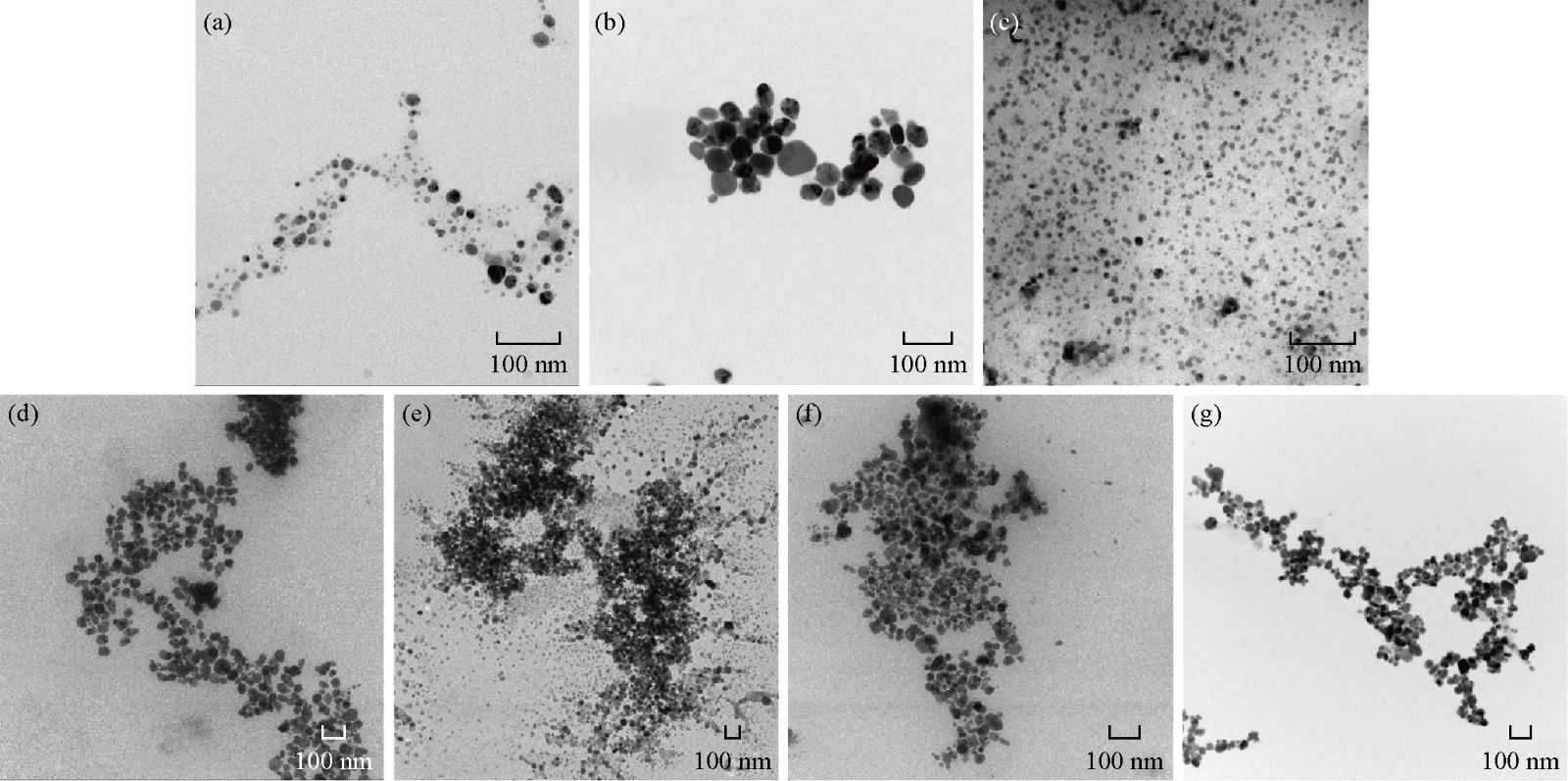
Fig. 4 TEM micrographs of nanoparticles and average size (AS): (a) PaNPs, AS: 14 ± 4 nm; (b) PeNPs, AS: 38 ± 12 nm; (c) PchNPs, AS: 11± 2 nm; (d) GsNPs, AS: 40 ± 6 nm; (e) RsNPs, AS: 40 ± 7 nm; (f) GfNPs, AS: 27 ± 5 nm; (g) BcNPs, AS: 34 ± 6 nm.
Colloidal stability assays
Due to their high resistance to centrifugation, nanoparticles obtained from P. atropurpurascens (PaNPs), P. expansum (PeNPs), P. chrysosporium (PchNPs) and G. spectabilis (GsNPs) were selected for their further characterization. All the selected nanoparticles were stable for five months when kept at 4ºC in 10mM buffer MES pH6, except GsNPs. The UV-Vis spectra ofthese nanoparticles showed that the original extinction peak decreased in intensity (due to the depletion of nanoparticles), and the nanoparticles aggregated at the long term. This indicated that in this case the organic shell surrounding the inorganic core was attached in a less stable way than that in the case of PaNPs, PeNPs and PchNPs. Thus, nanoparticles synthesized by these three fungi (P. atropurpurascens, P. expansum and P. chrysosporium) were selected for further characterization studies. Stability at different pH values (5 and 7) and ionic strength (10, 50 and 100 mM MES buffer) conditions of these purified nanoparticles were also evaluated. All nanoparticles were stable for most of the evaluated conditions (Fig. 5). However, a small decrease (15-20%) on the maximum absorbance intensity at the SPR was observed at 10 mM MES pH 5 for PchNPs and PeNPs, and at 100 mM MES pH7 for PaNPs. These differences in the range of pH, where they showed their highest colloidal stability, could be related with different compositions of the organic shell in terms of number of COOH and NH2 total groups.

Fig. 5 Stability at different pH (5 and 7) and ionic strength (10, 50 and 100 mM MES buffer) values of (a) PaNPs, (b) PeNPs, and (c) PchNPs.
DLS and -potential
The size distribution histogram of DLS indicated that the average sizes were 25nm, 30nm and 12 nm for PaNPs, PeNPs, and PchNPs. A comparison of average sizes of these three nanoparticles with the data obtained from the TEM analysis (Fig. 4) showed consistent results as PchNPs showed the smallest size (11 nm) and PeNPs showed the biggest size (38 nm) in TEM. Dispersity Parameter (PDI) showed a moderate polidisperse distribution (<0.4) from DLS results (Table 1). Besides, -potential determination showed that all nanoparticles had net negative surface charge at pH 6 (Table 1). The nanoparticles with highest net negative charge were PaNPs which were the most stable at acidic pH. This is in accordance with having an organic shell containing a larger amount of COOH groups.
Table 1 DLS and -potential of the nanoparticles
|
Nanoparticles |
DLS 1 |
-potential 2 (mV) |
|
|
Average Size (nm) |
PDI |
||
|
PaNPs |
25 |
0.2 |
-16.6 |
|
PeNps |
30 |
0.2 |
-11.5 |
|
PchNps |
12 |
0.3 |
-14.7 |
Note: 1Suspension of nanoparticles prepared with deionized water; 2The pH of the solution was 6.
Electrophoretic analysis
Agarose gel electrophoresis of nanoparticles is an interesting technique for their characterization as the mobility of the nanoparticles is sensitive to their size, charge and shape. Thus, after determining the nanoparticles with the best colloidal stability, they were loaded on a1% agarose gel. PaNPs, PeNPs and PchNPs showed a migration band towards the cathode as expected according to their good short and long-term colloidal stability, size and zeta potential analysis. Negatives values obtained for PaNPs, PeNPs and PchNPs in -potential measures are consistent with the results obtained in the electrophoretic analysis where at the pH of the running buffer (8) these nanoparticles showed negative charges on their surfaces. However, their relative mobility is different and it is in accordance to their observed net negative charge/size ratio. Thus, PchNPs with the smallest size (12 nm) and high net negative charge (-14.7 mV), has the highest ratio (1.2) and therefore the highest mobility. PeNPs, however, due to their largest size (30 nm) and lowest net charge (-11.5 mV) has the lowest ratio (0.4) and thus the lowest electrophoretic mobility (Fig. 6). RsNPs and BcNps were also loaded on the gel to show the importance of a good colloidal stability of the nanoparticles not only for their complete characterization but also thinking on their future application. In this case, as expected due to their low short-term colloidal stability these nanoparticles could not migrate due to their fast aggregation on the pH and ionic strength conditions of the electrophoretic buffer (45 mM Tris-Borate/1 mM EDTA, pH 8).

Fig. 6 Electrophoretic analysis in 1% agarose gel of BcNPs, RsNPs, PeNPs, PchNPs and PaNPs
Antimicrobial assays
Several studies have demonstrated that bactericidal properties of the silver nanoparticles are different from silver ions and they are strongly influenced by their shape, size, concentration, and colloidal state [18]. It has been found that reducing the size of nanoparticles enhances their stability and biocompatibility. In general, for nanoparticles to be effective, their size typically should be no larger than 50 nm, due to their higher penetration ability into bacteria, especially in Gram-negative bacteria [19]. The antimicrobial activity of PaNPs,PeNPs and PchNPs was evaluated through Agar Diffusion Method against Gram-positive and Gram-negative bacteria as they had an adequate size (10-30 nm) and excellent colloidal stability. All silver nanoparticles produced growth inhibition haloes against E. coli (Fig. 7(b) and (c)) but not against S. aureus, under the evaluated conditions. This difference could be attributed to the thicker peptidoglycan layer of Gram-positive bacteria, which protect the cell against several antibacterial agents. Similar results were previously reported [20], showing that the inhibition of S. aureus growth was less remarkable, while E. coli was inhibited at low silver nanoparticles concentrations. These results showed that the biogenic silver nanoparticles obtained had a potent antibacterial activity against E. coli, demonstrating their potential as effective antibacterial agents for their use in a diverse range of clinical and therapeutic applications.
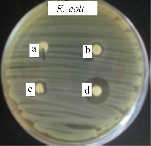
Fig. 7 Antimicrobial activity of PaNps against E. coli. (a)Extracellular extract; (b) and (c) PaNps (duplicate); (d) Gentamicine.
Conclusions
A comparative study about the biogenic synthesis of silver nanoparticles of twelve different fungal strains was carried out. As it could be observed in Fig. 8, a decision tree workflow was followed in order to select the best strains in terms of ability and yield of production, resistance to centrifugation and long-term colloidal stability of the obtained nanoparticles after their purification. Silver nanoparticles were synthetized with extracellular extracts from eight out of the twelve assayed fungi. Purification, characterization and colloidal stability assays were done. Due to a low yield of production (PoNPs), the impossibility of being purified by centrifugation (RsNPs, GfNpsand BcNPs) or a low long-term colloidal stability once purified (GsNPs), five out of eight of the producing strains were discarded. The nanoparticles showing the best characteristics (size, resistance to centrifugation, long-term colloidal stability once purified, and antimicrobial activity) were PaNPs, PeNPs and PchNPs. The sizes of these silver nanoparticles varied from 10 to 30 nm with a spherical shape. According to DLS and TEM measurements, PeNPs had larger size than PaNPs and PchNPs. This result was consistent with the lower migration electrophoretic mobility of these nanoparticles in the agarose gel. Negative values obtained for PaNPs, PeNPs and PchNPs in -potential measurements were consistent with the results obtained in the electrophoretic analysis where these nanoparticles showed negative charges on their surfaces. Besides, these nanoparticles were able to inhibit the growth of E. coli, demonstrating their potential for several biotechnological and biomedical applications.
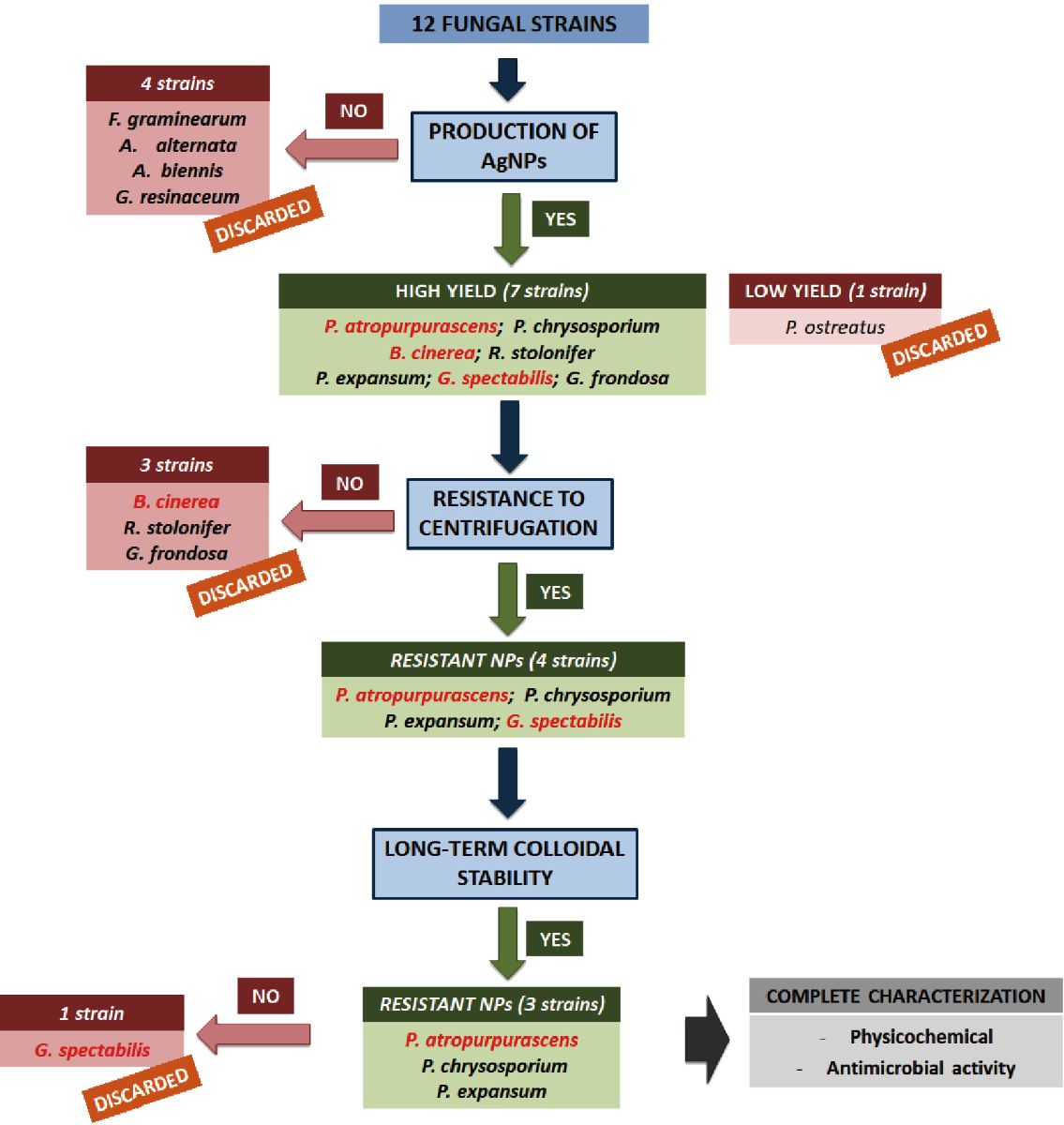
Fig. 8 Decision tree work-flow that allowed to select PaNPs, PeNPs and PchNPs as the best synthesized nanoparticles. Fungal species not previously reported as producing silver nanoparticles are shown in red.
Acknowledgments
This work was supported by the Comisión Sectorial de Investigación Científica (Universidad de la República, Uruguay), Program for the Development of Basic Sciences (PEDECIBA-Química) and Agencia Nacional de Investigación e Innovación (ANII), Uruguay. R. M. F. acknowledges financial support from MINECO (Ramón y Cajal grant RYC-2015-17640). The microscopy works were conducted in the Laboratorio de Microscopias Avanzadas, Instituto de Nanociencia de Aragon - Universidad de Zaragoza. Authors acknowledge the LMA-INA for offering access to their instruments and expertise.
Conflict of Interests
The authors declare that no competing interest exists.
References
Copyright© Paula Sanguiñedo, Raluca María Fratila, María Belén Estevez, Jesús Martínez de la Fuente, Valeria Grazú, and Silvana Alborés. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.