MIF-173G>C Polymorphism and Susceptibility to Cutaneous Leishmaniasis in Iraq
Hasan Raheem Khudhur *, Ghada Basil Alomashi
Department of Microbiology, College of Medicine, University of Al-Qadisiyah, Iraq.
* Corresponding author. E-mail: hassanraheem1990@gmail.com
Received: Apr. 16, 2018; Accepted: Jun. 7, 2018; Published: Jul. 16, 2018
Citation: Hasan Khudhur, Ghada Alomashi, MIF G-173C Polymorphism and Susceptibility to Cutaneous Leishmaniasis in Iraq. Nano Biomed. Eng., 2018, 10(3): 213-216.
DOI: 10.5101/nbe.v10i3.p213-216.
Abstract
Cutaneous leishmaniasis (CL) is a parasitic disease transmitted by biting of the sandfly; it is a severe health problem in many regions of the world. The disease is endemic in Iraq and other countries. In Iraq, there are two main species of the genus Leishmania causing the infection: L. tropica and L. major. Previous studies suggested that a genetic makeup of host also had an essential role in the outcome of the disease. The present study investigated the association between CL and functionally active polymorphisms in the macrophage migration inhibitory factor (MIF gene). Samples of peripheral blood were collected from 60 patients with CL and 32 apparently healthy controls. MIF-173G>C polymorphism was detected in patients and control groups by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique. There was no statistically significant difference in the MIF-173G>C polymorphism between the CL patients and the healthy controls (P = 0.234), as well as no association between MIF-173G>C polymorphisms and CL; the frequency of allele C showed a trend towards lower frequency in patients as compared to that observed in the control group (P = 0.512).
Keywords: Cutaneous leishmaniasis, Genetic susceptibility, MIF-173G>C polymorphisms
Introduction
Human leishmaniasis is a parasitic disease transmitted by sand flies. It is characterized by a spectrum of cutaneous, mucocutaneous and visceral diseases that depend largely on the species of the parasite involved and the host’s immune response [1, 2]. Cutaneous leishmaniasis (CL) is the most common form of leishmaniasis, about 1-1.5 million of cases, that is about 50 to 70% of all leishmaniasis cases every year in the world [2, 3]. Of all the CL cases each year, more than 90% occurred in five countries in the old world, that is Afghanistan, Algeria, Iran, Iraq and Saudi Arabia, and two countries in the new world including Brazil and Peru [4]. Leishmania major (L. major) and Leishmania tropica are considered as the common causes of CL in Iraq [5]. Macrophage migration inhibitory factor (MIF) is considered to be one of the first cytokines to be discovered, considered an essential component of the immune response of host against microbial and inducing activation and secretion of interleukins like TNF-α, IFN-γ, IL-1β, IL-12, IL-6 and IL-8 by immune cells [6]. MIF increases the survival of macrophage by inhibition activity of P53 and thus decreases the activation-induced apoptosis [7, 8]. Finally, cDNA was cloned in 1989 in humans, and MIF genomic localization to chromosome 22q11 was later mapped. The human MIF gene has three exons of 205, 173 and 183 bp, which are separated by two introns of 189 and 95 bp [6, 9]. Previous studies suggested that MIF played an essential role in the resistance of host to CL, and found human MIF activated the infected macrophage to kill L. major at a concentration of 1.5 -2.5 μg/mL in vitro [10]. Jesus et al. in Brazil found that MIF-173G>C polymorphism was associated with CL [11], suggesting that C allele induced lower levels of MIF cytokine in serum, and that the less secretion of MIF might bring about correlation between susceptibility and leishmaniasis. The present study aims to investigate the relation of MIF-173G>C polymorphism with susceptibility to CL infection in Iraqi population, specifically in AL-Muthanna Province.
Experimental
Subjects and study design
A total of 60 patients infected with CL selected from areas endemic for CL, including 34 males and 26 females, with mean age = 17 ± 13 years old. Patients were clinically diagnosed as patients with CL based on dermatologist diagnosis as well as clinical and parasitological parameters [12]. Meanwhile, the negative control group consisted of 18 males and 14 females, with mean age = 21 ± 9 years old, including 32 apparently healthy people without any cutaneous lesions and not suffering from any other diseases.
MIF-173G>C typing
Genomic DNA from blood samples was extracted by using Accupower®Genomic DNA extraction kit (whole blood). The extracted genomic DNA from blood samples was checked by using nanodrop spectrophotometer which checked and measured the purity of DNA through reading the absorbance at 260/280 nm. Genotyping for MIF-173G>C polymorphisms was performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) with two primers provided by Bioner Company, as described by Donn et al. [13], and the results are listed in Table 1. The volume of PCR mixture for polymorphisms was 50 μL, including pre master mix (Bioneer, Korea), 5 μL (20 ng/L) of genomic DNA, 2.5 L of each primer reverse and forward (10 pm/L) and volume complete with nuclease-free water. Thermocycler conditions were: 94 °C for 5 min, then 38 cycles of 94 °C for 30 sec, 59 °C for 30 sec, and 72 °C for 30 sec, followed by 5-min extension at 72 °C. Then PCR products were analyzed by agarose gel electrophoresis on a 1% agarose gel and stained with ethidium bromide (10 mg/mL). The PCR products were digested by using restriction enzyme Alu I and PCR- RFLP master mix according to company instructions (Biolabs, U.K.). Then, the PCR-RFLP product was analyzed by agarose gel electrophoresis (2.5%) and product analysis.
Table 1 Sequence of primers, restriction enzyme, and size of DNA fragments
|
Name |
Primer sequence |
R. enzyme |
Genotype |
Product size (bp) |
|
MIF-173G>C |
F: 5’-ACT-AAGAAA-GAC-CCG-AGG-C-3 |
Alu I |
GG |
268 and 98 |
|
R: 5’-GGG-GCA-CGT-TGG-TGTTTA-C-3 |
CC |
268, 205.98 and 63 |
||
|
CG |
205, 98 and 63 |
Notes: F = forward; R = reverse; R. enzyme = restriction enzyme
Statistical analysis
Statistical analysis was conducted by using SPSS 23. The determination of statistical differences among different groups and associations between allelic and genotypes of MIF-173G>C polymorphisms with susceptibility to CL was performed by using Pearson's chi-squared (ᵡ2) test and odds ratio with 95% confidence interval [14]. Probability of P ≤ 0.05 was considered to be statistically significant.
Results and Discussion
In total, 60 CL patients and 32 healthy controls were genotyped; equilibrium of Hardy-Weinberg was performed in both patients and controls. Fig. 1 and Table 2 show the distribution of MIF-173G>C genotypes and alleles among patients and controls that were detected by PCR-RFLP technique. At this locus there are three genotypes; GG of 268 and 98 pb, GC of 205/268/98 and 63 pb and CC with band sizes as 205/98 and 63 pb respectively. No significant difference in genotype distribution was found between CL patients and healthy controls (P = 0.234). The study referred to the high frequency of GG genotype (44/60, 73.30%) among patients when compared with (20/32, 62.5%) in healthy controls, P = 0.282. Moreover, there was no statistically significant result (P = 0.512) in G and C allele between patients and healthy controls. A high frequency of G allele in the patient group (102, 85%) was found in comparison with G allele (52, 81.25%) in the control group (P = 0.512), and lower frequency of C allele in the patient group (18, 15%) in comparison with that observed in G allele (12, 18.75%) in the control group (P = 0.512).
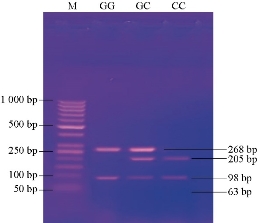
Fig. 1 Agarose gel electrophoresis image that shows the PCR-RELP product analysis of MIF gene by using Alu I estriction enzyme. M: Marker (1000-50 bp). Lane GG: Homozygote, the product digested with a restriction enzyme at 268 bp and 98 bp bands. Lane GC: Heterozygote, the product digested by restriction enzyme at 268 bp, 205bp, 98 bp bands and invisible at 63 bp band. Lane CC: Homozygote, the product digested by restriction enzyme at 205, 98 and 63 bp bands
Table 2 Distribution of MIF-173G>C genotypes and alleles
|
Variable |
Patients (60) |
Control (32) |
P value |
Odd ratio |
95% confidence interval |
Overall P |
|
|
Lower |
Upper |
||||||
|
GG |
44 (73.30%) |
20 (62.5%) |
0.282 |
1.650 |
0.660 |
4.125 |
0.234 |
|
GC |
14 (23.3%) |
12 (37.5%) |
0.151 |
0.507 |
0.200 |
1.289 |
|
|
CC |
2 (3.3%) |
0 (0%) |
-- |
-- |
-- |
-- |
|
|
Allele |
|
|
|
|
|
|
|
|
G |
102 (85%) |
52 (81.25%) |
0.512 |
1.308 |
0.586 |
2.920 |
0.512 |
|
C |
18 (15%) |
12 (18.75) |
0.512 |
0.765 |
0.343 |
1.707 |
|
This study investigated the association between CL and functionally active polymorphisms in MIF gene. Results of the detection of MIF-173G>C polymorphism among patients and controls were in agreement with the previous study [15]. In MIF-173G>C polymorphism, there was no significant difference in genotype distribution between CL patients and healthy controls (P = 0.234), and no significant difference (P = 0.512) in G and C alleles between patients and healthy controls. However, the current result suggested no association between MIF-173G>C polymorphisms and CL, which was in contrast with the previous study in Brazil having found a correlation between MIF-173G>C polymorphism and CL[11]. However, other potential factors should be taken into accounts, such as variant in the genetic makeup of their populations, the MIF-794CATT5-8 polymorphism, and the possibility of a correlation with different infecting Leishmania species, animal host species and vectors found in this area [16]. Therefore, all these factors may play essential roles in the susceptibility to CL.
Conclusions
In this study, no significant difference in genotype distribution between CL patients and healthy controls was found. It is suggested that MIF-173G>C polymorphisms do not affect susceptibility to CL in the sample subject from Iraq.
Conflict of Interests
The authors declare that no competing interest exists.
References
Copyright© Ghada Basil Alomashi, Hasan Raheem Khudhur. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.