Research Article
Application of Molecular Dynamics in Coating Ag-Conjugated Nanoparticles with Potential Therapeutic Applications
Farzin Sohraby 1, Mojtaba Hadi Soltanabad 1, Milad Bagheri 1, Masood Bezi Javan 2, Mostafa Javaheri Moghadam 1, Eisa Kohan Baghkheirati 3, Mohammad Bagher Bagherieh Najjar 1*
1 Department of Biology, Faculty of Science, Golestan University, Gorgan, Iran.
2 Department of Physics, Faculty of Sciences, Golestan University, Gorgan, Iran.
3 Department of Biology, Hakim Sabzevari University, Sabzevar, Iran.
* Corresponding author. E-mail: mb.bagherieh@gu.ac.ir Tel.: +98-17-32254161 Fax : +98-17-32245964
Received: Sep. 17, 2019; Accepted: Feb. 29, 2020; Published: Mar. 9, 2020
Citation: Farzin Sohraby, Mojtaba Hadi Soltanabad, Milad Bagheri, Masood Bezi Javan, Mostafa Javaheri Moghadam, Eisa Kohan Baghkheirati, and Mohammad Bagher Bagherieh Najjar, Application of Molecular Dynamics in Coating Ag-Conjugated Nanoparticles with Potential Therapeutic Applications. Nano Biomed. Eng., 2020, 12(1): 90-98.
Drug delivery systems may benefit from nanoparticles synthesized using biological methods. While chemical reduction of particles is facilitated by some active compounds present in the bio-extract, other active compounds, with potential therapeutic activities, may be adsorbed onto the surface of nanoparticles. However, the mechanism of bio-based nanoparticle synthesis is still under debate. Here, we first employed a molecular dynamics (MD) approach to theoretically predict the coating of a hypothetical 4.5 nm silver nanoparticle with four selected rosemary (Rosmarinus Officinalis L.) active compounds (rosmanol, isorosmanol, carnosol, and carnosic acid). Analysis of density maps and radial distribution functions (RDF) values suggested that the examined compounds had strong hydrophobic properties and could instantaneously be adsorbed to the nanoparticle surfaces. Next, we experimentally examined the capacity of rosemary leaf extract to synthesize and coat Ag-conjugated nanoparticles. The data obtained from ultraviolet–visible spectroscopy, transmission electron microscopy, Fourier-transform infrared spectroscopy and X-ray powder diffraction analyses confirmed the production of spherical Ag-conjugated nanoparticles with an average size of 12-15 nm, coated with proteins, secondary metabolites and other active compounds. Since this method can predict the dynamic behavior of therapeutic compounds when they are in contact with nanoparticles, we believe it provides a valid and new avenue to designing new therapeutic nanoparticles.
Keywords: Silver nanoparticle; Rosemary; Rosmarinus officinalis; Molecular Modelling; Molecular dynamics simulation.
Introduction
Drug delivery has long been a concern in pharmacology. Nanoparticles, with various shapes and sizes, have been proved suitable candidates for delivering hydrophobic drugs and biologics through biological barriers [1-3]. Synthesis of nanoparticles (NPs) using biological methods is considered to be safer, nontoxic, eco-friendly, and more affordable, as compared to physical or chemical methods [4-7]. Generally, in bio-based methods, natural compounds or extracts obtained from living organisms including bacteria, fungi and plants, containing chemical reducing activities, are employed in the synthesis of various NPs with different properties [8, 9]. Interestingly, during this biosynthesis reaction, several natural compounds present in the bio-extract can be readily attached onto the surface of those synthesized NPs. Subsequently, by changing solvent properties, such as pH, those compounds could be released from the surface of the NP. This phenomenon provides grounds for some interesting applications, including new means for isolation and purification of rare natural products, as well as new venues for drug delivery, using metallic NPs of choice, coated with selective therapeutic compounds. Several plant secondary metabolites, including phenolic compounds and alkaloids are thought to be able to act as an electron donor to metallic ions, converting them to neutral NPs [10-12]. Rosemary (Rosmarinus officinalis) is a well-known evergreen member of the Lamiaceae family, which contains several secondary metabolites with strong anti-platelet [13], antioxidant [14, 15], anti-inflammatory [16, 17], anti-diabetic [18], anticarcinogenic [16, 19], antibacterial, and antifungal activities [20, 21]. Among the various secondary metabolites present in rosemary leaf extract [22], carnosic acid, carnosol, rosmanol and isorosmanol are important diterpenes with potential valuable therapeutic applications [23, 24]; however, their properties and behavior in regard to attachment onto the surface of NPs have not yet been investigated. Molecular dynamics (MD) simulation is a widely used tool in computational biology, with great capabilities in revealing the underlying mechanisms of biological processes such as chemical and dynamic properties of biological macromolecules as well as their interactions with each other and their surroundings [25]. Here, for the first time, a molecular dynamics (MD) approach has been exploited to analyze and model the molecular behavior of four hydrophobic compounds present in rosemary leaf extract, in Ag-conjugated NP biosynthesis. Subsequently, the proposed model was evaluated by experimental biosynthesis and characterization of NPs coated with rosemary active compounds.
Experimental
Molecular dynamics simulation
In order to perform the molecular dynamics simulations, a 4.5 nm quasi-spherical silver NP, containing 3781 Ag atoms, kindly provided by Kyrychenko et al. [26], was placed in the center of a cubic box with a minimum distance of 1.5 nm from each side. Using this box, in total, five various systems were prepared and simulated, in four of which, 20 molecules of each active compound were inserted individually. In the fifth system, four active compounds were inserted altogether at a quarter (5 molecules each) of the concentration used in their individual systems. The molecules were inserted into the box at random positions with a concentration of approximately 57 mM. All MD simulations were performed by GROMACS 5.1.2 software [27] using gromos53a6 force field [28] and SPC water mode [29], under periodic boundary conditions (PBCs). The whole complex was solvated by TIP3P water molecules. The systems were energy-minimized, using the steepest descent minimization algorithm for all atoms [30]. Each system was equilibrated in both NVT ensemble (constant number of particles (N), volume (V), and temperature (T)) coupled to the V-rescale thermal bath at 300 K over 100 ps and in the NPT ensemble (constant number of particles (N), pressure (P), and temperature (T)) coupled to the Berendsen pressure bath at 1 atm over 300 ps. Each system was then subjected to a 30 ns molecular dynamics (MD) simulation under constant conditions of 1 atm and 300 K with a time step of 2 fs. Bond lengths constrained using the LINCS algorithm [31] and the long-range electrostatics were applied using the particle mesh Ewald (PME) [32], while the SETTLE algorithm [33] was employed to constrain the geometry of water molecules. The trajectory information was analyzed using GROMACS utilities and VMD programs [34]. Moreover, UCSF chimera was used for visualization, inspection and preparation of the NP and compounds [35]. The force field parameters of the compounds were obtained from the ATB web server for gromos53a6 force field and the Lennard Jones parameters and the partial charge of Ag atoms were obtained from already published data [26, 36, 37]. Although the actual partial charge of individual atoms in an Ag-conjugated NP molecule can dynamically vary between -0.07 and +0.07 [38], the net charge of the entire molecule is zero. To minimize the computing power needed for quantum mechanics calculations to an affordable value, the net- and partial-charges of the NPs were set to zero.
Biosynthesis and characterization of silver nanoparticles
The leaves of Rosmarinus Officinalis L. were collected from Golestan University campus, Gorgan, Iran (54°25'E; 36°50'N) during 2012-2013. Samples were washed and air dried at 60 ºC for 48 h and ground to a fine powder using an ordinary coffee grinder. One gram of each sample was used for extraction in 50 mL of ethanol for 48 h at room temperature. Afterwards, solutions were filtered through Whatman No. 3 paper and concentrated by a rotary evaporator at <40 °C. Silver-conjugated NPs were synthesized, as basically described by Hadi et al. [39]. 1% (V/V) of the plant extract and 1 mM of AgNO3 (Sigma-Aldrich, Germany) were mixed and retained at room temperature for 0, 45, 90, 150, 210 or 300 min. The solutions were then centrifuged at 20,000 g for 15 min and the resulting pellet was washed twice with distilled water and were left at room temperature for 16 h to dry before further characterizations. The progress of NP biosynthesis was monitored by measuring changes in the solution color, using an ultraviolet–visible spectrophotometer (Shimadzu UV-1800, japan) in the range of 200 to 800 nm. The crystallite structure of the particles was determined through recording their elemental spectra by an X-ray diffractometer (D8-Advance, Bruker, Germany) equipped with a CuKα radiation source (λ = 1.54 Aº) in the range of 20º to 80º at a 0.5 degree/s scan rate. The average crystallite size of the NPs was estimated by Sherrer's formula (D = 0.9λ/β Cosθ), where λ stands for the X-ray wavelength, β for the full width at half-maximum and θ for the diffraction angle. The data were analyzed with XPowder software package. A Philips CM120 transmission electron microscope was employed to determine the size and shape of the synthesized NPs, as previously explained [39]. FTIR (Fourier-transform infrared spectroscopy) was performed using a Spectrum RXI (PerkinElmer, USA) in the range of 4000–400 cm- 1 at a resolution of 4 cm-1, as described previously (40).The shape and size of the synthesized NPs were examined by a Philips CM120 transmission electron microscope at 120 kV with a 2.5 Angstrom resolution, as described by the supplier, as described previously [11].
Results and Discussion
Molecular dynamics simulation
The stability of the Ag-conjugated NPs was measured using the radius of gyration [41, 42], and is shown in Fig. 1. The radius of Ag NPs was slightly fluctuating while the Ag atoms vibrated in their positions during simulations. The stability of all NPs was declared by this method. The atomic motion and behavior of the compounds and their interactions with each other were studied by watching simulations’ trajectories [43-46]. Our analyses showed that each of the active compounds in question was instantaneously attached to the NP (Fig. 2 and Supplementary movie 1). As shown in Fig. 2, all twenty molecules of each compound present in the box could be attached to the surface of the NP, either directly or indirectly, suggesting that increasing the concentration of the molecules present in the box resulted in the attachment of all compounds onto the NP until the entire surface was occupied. The above-mentioned rosemary’s four active compounds (isorosmanol, carnosol rosmanol and carnosic Acid) have hydrophobic properties; i.e. they try to escape from interaction with the polar water molecules. This may explain why they immediately attached onto the NP surface or other compounds present in the solution (Supplementary movies 2 and 3). These four compounds were very similar in structure, and while all had functional hydroxyl and carbonyl groups, the three hydrophobic rings present in their structures dominantly made the compound’s molecular behavior similar to a hydrophobic molecule, which resulted in the immediate interactions of rosemary’s active compounds with each other and NPs. In order to analyze the movements of active compounds relative to the target NP, the density map of ligands during the simulation process was calculated by utilizing the Gromacs densmap program. Gromacs densmap produced a 2D density plot of a selected group across the box (Fig. 3), which shows the presence-time of a chosen group in a specific location using color intensity (i.e. darker regions are indicative of longer presence). For clarity, we manually and schematically placed the Ag-conjugated NP on the density map. All four active compounds were attracted to the Ag NP, and maintained their positions during the simulation period (Fig. 3). In addition, radial distribution functions (RDFs) were used to analyze the tendency of the active compounds to attach onto the Ag-conjugated NP surface. As shown in Fig. 4, the active compounds had the highest distribution density at 1 nm distance from the NP, which confirmed the attachment of all present onto the surface of Ag NP [47, 48]. In order to learn about possible key interactions influencing the attraction of active compounds to the NP surface, five separate MD simulations were performed. The results of these experiments showed that when the active compounds were approaching the NP, and their orientations were only affected by their initial orientation, while most of the molecules interacted with their hydrophobic rings [49] (Supplementary movies 4 and 5). In MD simulations, assigning the accurate atomic partial charge of each atom is a key step to correctly modelling a molecular system behavior. Several methods have recently been developed to calculate the atomic partial charges of a molecule,however, none are adequately qualified for large molecules or complex structures like NPs. Solving quantum mechanics equations even for a single Ag atom needs ample computer resources. Considering that an Ag-conjugated NP contains thousands of atoms, it is almost impossible to accurately calculate its partial charges even by advanced computers [50]. Therefore, we set the atomic partial charges of NP to zero, which provides an approximate model of Ag NP in hand [51, 52].
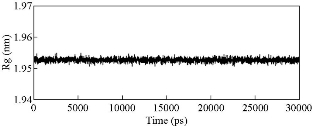
Fig. 1 The radius of gyration of Ag nanoparticle in one of the simulation systems.
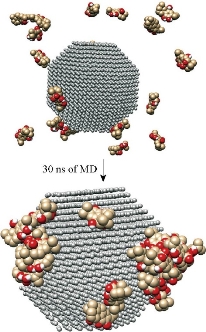
Fig. 2 The surface view of the Ag-conjugated nanoparticle after 30 ns of MD simulation. All of the 20 molecules could be absorbed on the surface of the nanoparticle.
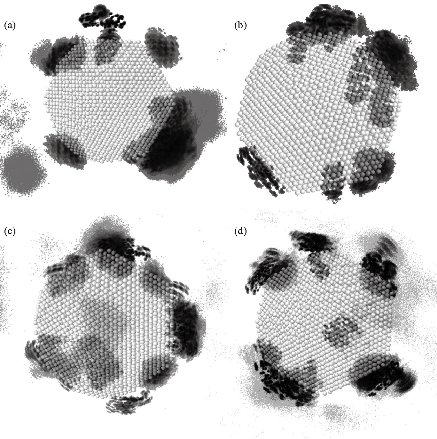
Fig. 3 Density maps of the four active compounds throughout the 30 ns simulation: (a) Isorosmanol, (b) carnosol, (c) rosmanol, and (d) carnosic acid.
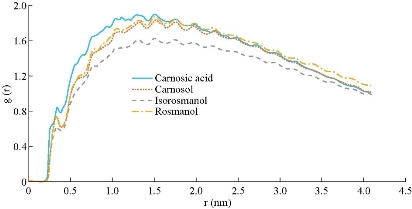
Fig. 4 The radial distribution function of the four active compounds during 30 ns simulation.
Experimental synthesis of silver nanoparticles
Chemical reduction of Ag+ to Ag-NPs during exposure to 1% ethanolic extract of Rosmarinus officinalis L. characterized by the color change of solution from yellow to dark brown after addition of AgNO3 is presented in Fig. 5. The color changes are basically caused by a quantum phenomenon that occurs in metallic NPs called excitation of surface plasmon resonance [53, 54]. The exact mechanism of biological reduction of Ag+ to Ag is not completely understood. The reduction of Ag+ can happen via reducing agents present in plant extracts such as proteins, secondary metabolites and biologically active compounds, which can also coat the Ag-conjugated NPs and prevent their aggregation. Recently, synthesized stable monodisperse Ag-protein (core–shell) NPs showed an effective antimicrobial potency against two representative bacteria, Staphylococcus aureus and Klebsiella pneumonia. Proteins bind to the NPs either through their free amine groups or cysteine residues [55, 56]. As it was demonstrated in MD simulation, active compounds and secondary metabolites, can bind to the NPs too. The transmission electron microscopy (TEM) images showed that the size of the synthesized NPs varied between 3 to 30 nm in diameter, with an average size of 12-15 nm and a spherical shape (Fig. 6).
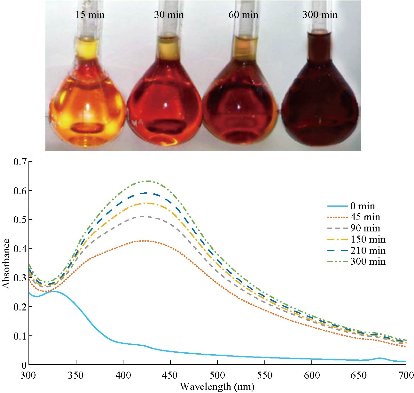
Fig. 5 The ultraviolet-visible spectra of Ag-NPs synthesized by the reduction of silver ions using rosemary leaf extract. The color change of reaction mixtures is depicted on the graph.
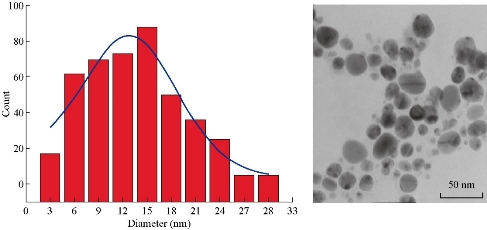
Fig. 6 The transmission electron microscopy image of the synthesized Ag nanoparticles with an average diameter of 12 to 15 nm.
Fourier-transform infrared spectroscopy
Fourier-transform infrared spectroscopy (FTIR) of the biosynthesized Ag-conjugated NPs by rosemary leaf extract suggested that several active compounds present in the extract were attached onto the surface of the synthesized NPs (Fig. 7(a) and Table 1). In the FTIR spectrum, several strong, broad, sharp and weak peaks were observed. The broad and strong peak at 3433 cm-1 represented O-H bond stretching in alcohols and phenols which corresponded to the O-H and phenol groups in the rosemary’s active compounds. The 2340 cm-1 strong and sharp peak might probably be related to CO2 contamination. The sharp and weak peak at 1610 cm-1 might be related to the C-H bond in the aromatic stretching, associated with the phenolic ring structures or it could be related to aromatic rings which might be indicative of the aromatic amino acids of various proteins, or the aromatic rings of the rosemary’s diterpenes. The FTIR spectra also indicated the non-bonded chemical interactions between rosemary leaf extracts and Ag-conjugated NPs. These observations suggest that several rosemary’s metabolites may act as bio-reductants and/or bio-stabilizers. Moreover, the NPs coated with these metabolites may exhibit unique therapeutic properties.
Table 1 The details of the frequencies and the corresponding bonds
|
Absorption (cm-1) |
Group |
Compound class |
Appearance |
|
3433 |
O-H stretching |
alcohol, phenol |
strong, broad |
|
2340 |
O=C=O stretching |
carbon dioxide |
strong, sharp |
|
1610 |
C=C stretching |
cyclic alkene |
medium |
|
1433 |
O-H bending |
carboxylic acid |
medium |
X-ray powder diffraction analysis
In order to understand the nature and composition of the synthesized Ag-conjugated NPs, X-ray powder diffraction (XRD) analysis was performed, which is indicative of the presence of different atoms in the solution based on the observed peak patterns (Fig. 7(b)). The (111), (200), (220), (222) and (311) peaks represented the AgCl molecules in the solution and the (111) and (200) peaks represented Ag atoms [57], suggesting that in addition to Ag-NPs, AgCl-NPs were present in the solution (Fig. 7(b)). Similar results were reported when other bio-based NP biosynthesis methods were employed [57, 58].
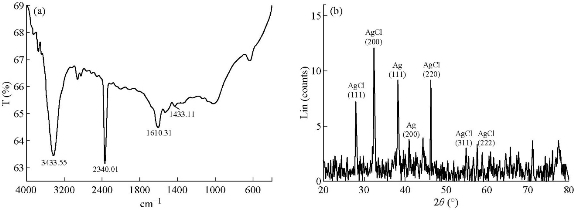
Fig. 7 (a) The Fourier-transform infrared spectrum of synthesized Ag/AgCl–NPs. (b) X-ray powder diffraction pattern of Ag/AgCl NPs which represented the presence of both Ag and AgCl atoms in the solution.
Conclusions
Recently, NPs have immensely influenced human lives, especially considering their applications in drug delivery. Data obtained from molecular dynamics simulation of the coating process of Ag-conjugated NP by rosemary’s active compounds, revealed that rosmanol, isorosmanol, carnosol, and carnosic acid were able to successfully attach to the surface of the synthesized NPs, probably due to their hydrophobic properties. The in-silico biosynthesis modelling was confirmed by experimental data, and indicated that the synthesized NPs consisted of Ag/AgCl with a spherical shape and an average size of 12-15 nm, coated with several bioactive compounds of rosemary leaf extract. Therefore, a new idea is presented for taking advantage of plants’ abundancy for biosynthesis of NPs coated with desired active compounds, with promising beneficial applications in future drug discovery approaches.
Acknowledgements
The authors are very grateful to Stephen Cork (Charles Sturt University, Australia) for providing constructive comments during manuscript preparation. This study was funded by Golestan University.
Conflict of Interests
The authors declare that no competing interest exists.
References