Research Article
In-vitro Study on Genotoxicity of Green Synthesized Silver Nanoparticles
Marcellin Mecwan 1, Manita Das 2, Sonal Thakore 2, Sonal Rajiv Bakshi 1*
1 Institute of Science, Nirma University, Ahmedabad, Gujarat 382481, India.
2 Department of Chemistry, The Maharaja Sayajirao University of Baroda, Vadodara, India.
* Corresponding author. E-mail: sonal.bakshi@nirmauni.ac.in, Tel.: 079-71652755, Mobile: +91 9426417807
Received: Aug. 13, 2020; Accepted: Jan. 18, 2021; Published: Feb. 19, 2021
Citation: Marcellin Mecwan, Manita Das, Sonal Thakore, and Sonal Rajiv Bakshi, In-vitro Study on Genotoxicity of Green Synthesized Silver Nanoparticles. Nano Biomed. Eng., 2021, 13(1): 72-81.
DOI: 10.5101/nbe.v13i1.p72-81.
Abstract
The wide use of metal nanoparticles has raised the concern of their release in the environment and their biological safety. The use of silver nanoparticles (Ag NPs) as effective antimicrobial agent is also questioned due to the resistance developed by pathogens and the biosafety for the host. The genotoxicity caused by such particles need to be assessed to ensure that they are safe to use. In this study, green synthesized Ag NPs were used and the biosafety in terms of genotoxicity was evaluated by in vitro Chromosome aberration (CA) assay using short-term cultures of human peripheral blood. The physicochemical parameters of these Ag NPs were studied for stability in nano form. The transmission electron microscopy (TEM) and energy-dispersive spectroscopy (EDS) were carried out. The dynamic light scattering and zeta potential measurements revealed stable nano particles. The UV visible & fluorescence spectroscopy for binding affinity of Ag NPs did not suggest significant DNA interaction. The exposure of whole blood cultures to Ag NPs for 24 h showed dose dependent increase in frequency of CA per cell at 350, 650 and 1000 µL (P = NS, P < 0.05 and P < 0.001 respectively). Our study demonstrates that genotoxicity of Ag NPs can be reduced by the use of green synthesized Ag NPs at low dose exposure, which will guide selecting right concentrations of NP for further in vivo studies, and future applications.
Keywords: Ag NPs, Green synthesis, Characterization, DNA binding, Genotoxicity, Chromosomal aberrations
Introduction
Nanoparticles (NPs) are widely used due to their unique properties in several fields such as electronics, cosmetics, paintings, and packaging as well as in industrial, domestic, and biomedical products. The intended and unintended exposure to NPs of environment and humans raises concern due to their prolonged sustenance and potentially higher concentrations by various routes. The exposure may occur via three main portals of entry: (1) Inhalation via the respiratory tract, (2) Dermal exposure via skin and (3) Oral exposure via the gastrointestinal tract [1, 2]. Like any xenobiotic, NPs can cause cellular toxicity in short term in addition to various possible detrimental effects on long term. The nanoparticles can be classified into metal NPs, carbon based NPs, ceramic NPs, polymeric NPs and semiconductor NPs. The metal NPs are focus of interest because of their wide use and potential in nanotechnology, mainly the silver nanoparticles (Ag NPs). The Ag NPs have anti-microbial properties and hence are attractive alternative to antibiotics, the wide use of which has led to the microbial resistance in bacteria. Few studies have reported the mechanism of bacterial resistance to Ag NPs [3, 4] while Panacek et al. reported development of resistance to Ag NPs after repeated long-term exposure in bacteria [5]. Several strategies of nanoparticles, especially Ag NPs to fight multidrug resistant bacteria was described by Baptista et al. [6]. One approach could be the use of plant extract for the synthesis of Ag NPs. When these nano sized metallic particles adapted for the synthesis, they can considerably change physical, chemical & biological properties due to their increased surface-to-volume ratio [7, 8], and thus the bottom-up approach [9] is employed due to its feasibility and less toxic effects to design monodispersed and stable nanoparticles. Physicochemical properties of NPs such as size, shape, surface charge, chemical composition, agglomeration and crystal structure which influence the toxicity of NPs, need to be assessed while considering toxicity study. The size, zeta potential and TEM and EDS analysis of Ag NPs were carried out in our study. For the synthesis of stable nanoparticles, the conventional physical and chemical methods are used which can be very expensive and hazardous as it often involves high temperature, pressure and use of harmful chemicals for the reaction. Hence, the use of biological methods are most preferable as it shows high stability, high yield and solubility at a lesser cost [8, 10]. Green synthesis of NPs by biological methods using micro-organisms, enzymes, plant extract and panchakavya [11] are reported to be eco-friendly. A complete green synthesis of Ag NPs following green-chemistry principles was first reported by Raveendran et al. [12]. Thereafter there have been several reports of the various methods of green synthesis of Ag NPs in which many included the use of plant extracts [13]. The biosynthesized Ag NPs produced by the leaf extract of Hydnocarpus pentandra found to be quite stable in solution and showed considerable antioxidant and excellent anticancer activity against breast cancer cells (MCF7), however, further in vivo studies are need to be carry out for considering green synthesized Ag NPs as alternative medicine for cancer therapy [14]. The study by He et al. demonstrated for the first time the antitumor activity of green synthesized Ag NPs against human lung cancer in vitro and in vivo [15]. Their result concluded the strong inhibitory effect of Ag NPs on the growth of human lung cancer H1299 cells as well as a potently suppression of growth on a mouse H1299 xenograft tumor model. More reports comprise the use of plant extracts like Cinnamon zeylanicum [16], Stevia rebaudiana [17], Euphorbeace latex [18], Cassia angustifolia [19], Ocimum tenuiflorum [20], carob [21], sunflower oil [22], papaya leaf extract [23] and seed extract of Derris trifoliate [24]. Previous study by Otunola et al. reported biosynthesis of spice blend Ag NPs for the first time with high antibacterial and antioxidant activities of Ag NPs against selected bacterial pathogen and free radicals, respectively [25]. In the present study, Ag NPs were synthesized using cinnamon cassia extract as a capping agent. The cinnamon extract contain sugars, terpenoids, polyphenols, alkaloids, phenolic acid and protein which are excellent reducing agents in synthesis of NPs, they are mainly responsible for the conversion of Ag+ to Agо and this bio-reduction of metal ions imparts stability of the size and morphology of NPs [26, 27]. The increased use of NPs have raised concerns for biological safety due to the longer sustenance of NPs and their reactive nature. Most severe outcome of biological toxicity is when a chemical agent reacts with the nuclear material and causes genetic damage which increases the risk of cancer, mutations in gametes and thus abnormal or loss of progeny. It is crucial to identify the cause through the detection of DNA lesions, chromosomal damage or recombination. The standard methodologies of in vitro genotoxicity testing need to adapt when working with NPs due to their unique properties. Ag NP induced DNA damage (DNA double-strand breaks) and chromosomal aberrations such as acentric and dicentric chromosomes, chromosomal fusions and fragments are reported [28-30]. Among many reasons of Ag NP genotoxicity, one include the toxic effect produce by Ag NPs due to oxidative stress while their interactions with cell membrane [31]. Several reports have mention that the release of Ag+ ions may have influence on Ag NPs genotoxicity; some studied that their role is negligible, while others obtained indirect or inconclusive evidence of their influence on the Ag NPs genotoxicity [32-35]. A few studies have reported the amount of ROS generation may enhanced by intracellular Ag NPs [36, 37] which cause multiple intracellular effects, including DNA damage leading to mutagenesis [38]. Asharani et al. [30] and Mcshan et al. [38] have reported that Ag NPs can also interact with mitochondria, disrupt the electron transport chain and cause production of ROS, interrupting ATP synthesis; increase in ROS and reduction of ATP result in protein and DNA damage. ROS are considered to be great source of spontaneous damage to DNA. Ahamed et al. has examined DNA damage response to polysaccharide surface functionalized (coated) and non-functionalized (uncoated) Ag NPs in two types of mammalian cells & observed different surface chemistry of Ag NPs induce different damage response [28]. This investigation may correlated to long-term effects of mutagenesis or carcinogenesis. The results by P. V Asharani et al (2010) demonstrated Ag NPs effect on human erythrocytes and concluded that Ag NPs in general have haemolytic and haemoagglutinating potential suggesting genotoxicity of particles and could trigger development defects in embryos [39]. Moreover, Ghosh et al. stated apoptosis and DNA strand breaks in human lymphocytes revealed that Ag NPs at concentration 25 µg/mL can cause genotoxicity [40]. More Ag NPs genotoxicity data are reported in the review by Rodriguez-garraus et al. [31]. The present study aimed to assess the potential genotoxicity of Ag NPs in vitro using short term cultured human blood lymphocytes in terms of induced levels of chromosomal aberrations (CA) following treatment with NPs as compared to controls [41]. The results of CA assay indicate genotoxicity of NPs which can be by DNA binding or by increased oxidative stress in cellular microenvironment respectively. The study of DNA interaction with Ag NPs was assessed by fluorescence absorbance in addition to CA assay for in vitro cytogenetic damage.
Experimental
Materials/chemicals
Silver nitrate (AgNO3) was procured from Merck (New Delhi, India) and cinnamon was purchased from the local market. Sodium citrate, sodium chloride, methanol and glacial acetic acid of analytic grade were obtained from Merck (Mumbai, India). Complete growth medium; RPMI-1640 HiKaryoXLTM-AL165A; low melting agarose, DMSO, EDTA and Ethidium bromide, were purchased from Hi-Media (Mumbai, India). Human genomic DNA was isolated from normal peripheral blood lymphocytes as per the manufacturer’s protocol (Qiagen midi kit, Cat. No.51104).
Biosynthesis of Ag NPs
Preparation of Cinnamon extract
Cinnamon cassia was used as a stabilizing agent, which prevents the agglomeration of NPs. The extract was produced by adding 2000 mg of cinnamon powder into 100 mL of deionized water (solvent medium), sonicated for 30 minutes and kept on stirring for overnight followed by the filtration with Whatmann filter paper no. 40. The final extract was stored at 4°C.
Preparation of reducing agent
Sodium citrate was used as a reducing agent that prevents the metallic form to turn into ionic form. Reducing agent was prepared by adding 1 gm of Trisodium citrate dihydrate (10%) into 10 mL of deionized water.
Preparation of 0.01M AgNO3 solution:
AgNO3 0.0084g was added into 5 mL of deionized water, mixed thoroughly and stored at 4-6°C in amber glass flask to prevent auto oxidation of Ag.
Synthesis of Silver nanoparticles using Cinnamon extract
Ag NPs were obtained by the stirring 50 mL of Cinnamon extract for 10-15 minutes and adding 1 mL of AgNO3, and further stirring for 10-15 minutes. 2.5 mL of reducing agent was added and alternate treatment of stirring and heating was provided till the color of the solution changed from yellow to brown. The reason of color change is due to the reduction of nitrate from Ag+ to free reduced form of Ag which indicate synthesis of Ag NPs.
Nanoparticle characterization
Dynamic light scattering method was employed for the zeta potential and particle size analysis was done by Malvern Zetasizer (Nano ZS, Malvern Instruments Ltd., Malvern, UK). For TEM measurements, a drop of synthesized Ag NPs was placed under lyophilization before loading on to a specimen holder. FEI (Technai G2, F30) electron microscope was used at an acceleration voltage of 300 kV to carry out High-Resolution Transmission Electron Microscopy (HR-TEM) analysis of the sample. Energy dispersive X-ray (EDX) analysis of the sample was recorded by the model-JSM-5610 LV attached to Scanning electron microscopy (SEM).
Isolation of genomic DNA from blood (As per QIAamp DNA blood midi kit)
Human genomic DNA was isolated from whole blood collected from a healthy donor as per the manufacturer’s protocol of QIAamp DNA blood midi kit.
DNA binding study
UV-Vis measurements using PBS buffer
UV absorption spectra of human genomic DNA suspension (3.33 µg/mL, total: 10 µg) was recorded from 230-300 nm following addition of increasing volumes of Ag NPs (0–82 μL). Experiment was carried out in 1x phosphate buffered saline (PBS), pH 7.4, in a conventional quartz cell attached with a thermostat to maintain the temperature at 25°C. Mixture was incubated for 2-3 minutes upon adding an increasing volume of Ag NPs to the mixture to attain the set temperature (25°C) before each measurement. The absorbance profiles were recorded using Carry-60 UV visible spectrophotometer.
Fluorescence measurements:
Fluorescence measurements were performed on a Cary Eclipse fluorescence Spectrophotometer (Agilent Technologies, Richardson, TX, USA) by using fluorescence cuvette having 1cm path length for all experiments and measurements were recorded at 25 ˚C. EtBr was used as fluorescence stain. Intercalation of EtBr between the base pairs of DNA leads to significant increase in fluorescence. 20 µL sample of human DNA in 2965 µL deionized water containing 15 µL EtBr was titrated with increasing concentrations of Ag NPs and the mixture was kept for 2- 3 minutes before each measurement. Manual addition of Ag NPs was carried out in a series of 2, 12, 22, 32, 42 and 52 µL. Fluorescence emission spectra were recorded at 605nm excitation wavelength and 570-650 nm emission wavelengths. The fluorescence quenching data were analyzed according to the Stern-Volmer equation 1, F0/F= 1 + KSV [Q], where F0 and F are the fluorescence intensities in the absence and presence of different concentrations of quencher [Q] Ag NPs, respectively. KSV is the Stern-Volmer quenching constant, which was obtained from the slope of the plots F0/F versus [Q] [42].
Short-term blood culture for chromosome aberration assay
Human peripheral blood lymphocytes in the whole blood of healthy individuals was used as a test system in this study after approval of the institutional ethical committee. The blood donor was selected after informed consent and taking detailed history to ensure healthy status and excluding recent past exposure to drug, radiation, and infection etc. known confounding factors. The aseptically collected blood in heparinized vial was used for short term cultures. 1.0 mL blood was cultured in 10 mL of RPMI-1640 complete growth medium and incubated at 37 ℃ for 48 h. At the 48th hour, cultures were exposed to 350, 650 and 1000 µL Ag NPs; Mitomycin-C (positive control), and 1000 µL cinnamon extract (vehicle control). One tube was kept as it is (without any addition) as a negative control. Colchicine (0.5 µg/mL) was added at 70th hour and cells were harvested at the 72nd hour by hypotonic treatment (0.56% KCl, 20 min) and fixed with Carnoy’s fixative 3:1 (methanol: glacial acetic acid) with repeated centrifugation (at 3000 rpm) and multiple changes till clear pellet was obtained. Slides were prepared by air-dry method and stained with 4% Giemsa for 6–8 min in Sorensen buffer (pH 7.0). Slides were coded before microscopic analysis and at least 100 well spread metaphases were scored per experiment (100 cells/replicate culture) while recording structural chromatid and chromosome aberrations (gaps and breaks). The scoring criteria for chromosomal aberration were as described by the OECD guidelines [41].
Result and Discussion
Silver nanoparticles characterization by Zeta sizer
The Zeta sizer Nano range instrument is used to measure three parameters such as particle size, zeta potential and molecular weight. In zeta sizer, light passes from the three different angles and give three distinct sizes of NPs. Out of three different measurements if one dimension is between 1-100nm, the particles are considered NPs. We have obtained sizes of NPs as 67.6nm, 100.9nm and 159.4nm from three separate angles of zeta sizer (Fig. 1). Zeta potential: The electric potential at the boundary of the double layer is known as the Zeta potential of the particles and has values that typically range from +100 mV to -100 mV. Nanoparticles with Zeta Potential values greater than +25 mV or less than -25 mV typically have high degrees of stability. Dispersions with a low zeta potential value will eventually aggregate due to Van Der Waal inter-particles attractions. In this study the Zeta potential (Mean) was -9.1mV and it is between +25mV to -25mV which indicated that Ag NPs were in a stable form which is due to repulsions existing between particles. Fig. 2 shows the electrophoretic mobility of the Ag NPs was -0.000070 cm2/Vs.
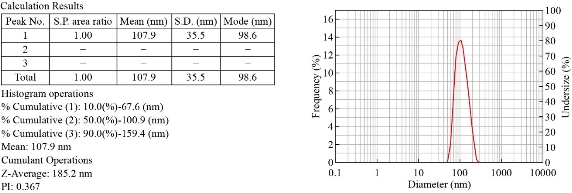
Fig. 1 Results of size distribution analysis of Ag NPs using DLS.
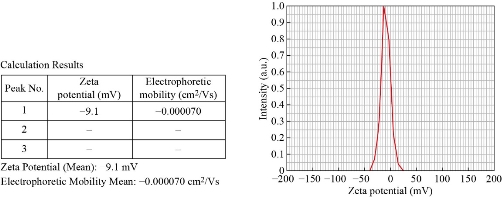
Fig. 2 Results of Zeta potential analysis of Ag NPs.
EDS and TEM analysis
EDS spectrum of biologically synthesized Ag NPs revealed the presence of elemental silver in the sample (Fig. 3). The graph plot depicts ionizing energy (keV) versus intensity of X-rays (counts). The strong signal at 3 keV denoted the existence of metallic silver. The reduction of silver ions to elemental silver thus confirms the formation of Ag NPs. The peaks of C, N and O are also arisen due to the formation of capping layer on the surface of nanoparticles by cinnamon extract. Quantitative analysis elucidate that C, N, O and Ag elements exhibited weight percentages of 54.65%, 11.20%, 21.31% and 12.84% respectively (Fig. 3). The morphology, dimension and distribution of the synthesized nanoparticles’ were visualized using transmission electron microscopic analysis. Images clearly displayed the nanoparticles in a range of magnifications indicating that the synthesized Ag NPs are spherical in shape and are in a dispersed form (Fig. 4).
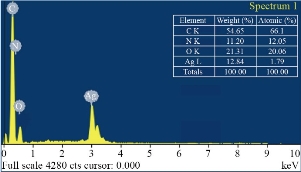
Fig. 3 Energy-dispersive spectroscopy (EDS) of Ag NPs and elemental analysis.

Fig. 4 TEM images (a,b) of Ag NPs at different magnification and (c) HR-TEM of silver nanoparticles.
DNA binding assay
Human genomic DNA isolated from whole blood was quantified by Nanodrop (Jenway, Geneva Nano) that revealed 85.0 ng/µL concentration with ratio of 1.9 (260/280 nm).
UV-visible spectroscopy
The interaction of Ag NPs with human genomic DNA was analyzed using UV absorption spectroscopy technique in order to provide evidence for the possibility of Ag NPs’ binding to human genomic DNA. The absorption spectra of the DNA-Ag NPs interaction have been recorded at a constant DNA concentration for different Ag NPs concentrations (0–82 μL) shown in Fig. 5. Generally, as absorption spectroscopy helps to determine characteristics of metal complexes with DNA, the spectral changes of DNA are observed in the form of hyperchromism and hypochromism concerning its double-helix structure. Hyperchromism suggests breakage of secondary structure of DNA which was reported by Rahban et al (2010) in the spectroscopic studies of Ag NPs with calf thymus DNA at 260 nm. Their work reported the interaction of Ag NPs with only CT DNA, where the DNA concentration was fixed and the changes in the absorbance of DNA around 260 nm with the addition of Ag NPs were monitered, which resulted in hyperchromic effect (increase in absorption) [43]. The study done by Pramanik et al (2016) have fixed the Ag NPs concentration and monitored the changes in the plasmonic absorption of Ag around 400 nm with addition of DNA, on the other hand, reported hypochromic effect (reduction in intensity). Further, Pramanik et al (2016) also observed the change in the absorbance of DNA at 260 nm by the addition of increasing amount of Ag NPs to the fixed concentration of DNA, similar to what has been reported by Rahban et al (2010). Moreover, their study to understand the specificity between double stranded and single stranded DNAs revealed strong interaction of ds DNA with Ag NPs compared to ss DNA [44]. The metal complex binds with the DNA via covalent and or/non-covalent interactions. Covalent interactions are via alkylation or inter and intra strand crosslinking [45] whereas non-covalent interactions include (1) intercalation between the base pairs, (2) binding to the major groove, minor groove or sugar phosphate backbone and (3) binding to the exterior of the helix through non-specific interactions [46]. All in all, if the metal complex binds with the DNA, it must show spectral changes by an increase or decrease in absorbance. The nitrogenous bases in nucleotides have strong optical absorption maxima around 260-280 nm. The interaction between NPs and DNA is examined by the changes in absorbance maxima of DNA, which was in the same range (260- 280nm), suggesting no significant Ag NPs interaction with the DNA in our study.
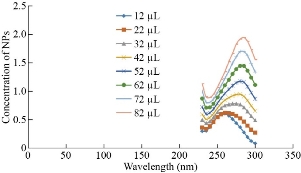
Fig. 5 UV-Vis absorbance spectra of human genomic DNA in the presence of different concentration of Ag NPs at 230-300 nm.
Fluorescence spectroscopy
The fluorescent DNA stain Ethidium Bromide (EtBr) binds with DNA to give fluorescence which is reduced when there is competition for DNA binding by another molecule like NPs indicating displacement of fluorescent dye. EtBr consisting of a planar phenenthridinium ring strongly fluoresces in presence of DNA due to intercalation between the adjacent DNA base pairs. The extent of fluorescence quenching of EtBr bound to human genomic DNA can be used to determine the extent of binding of second molecule with human genomic DNA. In this study on addition of Ag NPs there was no significant decrease in the fluorescence suggesting no significant interaction of synthesized Ag NPs with DNA (Fig. 6) whereas we have earlier reported fluorescence decrease with increasing concentration of TiO2 NPs [1, 47].
![]()
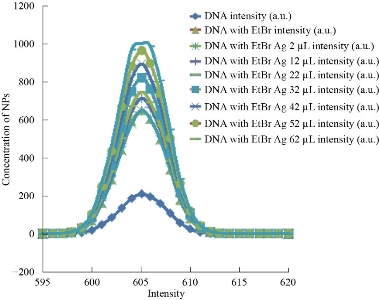
Fig. 6 Fluorescence emission spectra of intercalated Ethidium bromide and human genomic DNA incubated with increasing concentration of Ag NPs at 570-650 nm.
Chromosomal aberration assay
The peripheral blood lymphocyte cultures were exposed to Ag NPs at the 48th hour, and harvested at the 72nd hour. Considering the average cell cycle duration 18 hours, NPs were added at 48th hour after culture initiation, when cells divide asynchronously. After the addition of NPs, cells need to undergo S phase, to express aberrations. Since asymmetrical structural chromosome aberrations prevent unlimited division, harvesting cells when they are in first division post exposure of test substance is crucial. A harvesting time of 1.5 cell cycles is considered to be optimum for detecting clastogens as at the time of harvesting, most cells will be in their first cell division after exposure to NPs. The type of structural chromosome aberrations observed at metaphase reflects the duplication status of chromosomes in the treated cell. The genotoxic potential of Ag NPs was assessed by short term human peripheral blood lymphocytes following exposure for 24 hours. A minimum of 100 metaphases per sample per replicate were scored for the Chromosomal aberration assay. Fig. 7 shows the mean no. of chromosomal aberrations per cell when exposed to 350, 650 and 1000 µL Ag NPs, as compared to untreated cultures and treatment with the vehicle control in order to quantify the induced frequency of chromosomal aberrations The graph indicates that chromosome aberrations increase in a dose dependent manner as a result of exposure to Ag NPs as compared to untreated cultures with statistical significance except in case of lower concentration i.e. 350 µL. The non-significant increase in CA/cell value following exposure to vehicle control indicates the effect is due to Ag NPs only. The positive control showed highly significant increase in CA/cell value as compared to negative control which confirms that data obtained for Ag NPs is under appropriate experimental conditions. A report by Patel et al. mentioned significant increase in chromosome aberration with 75 and 125 µM TiO2 NPs whereas no significant difference at lower concentration (25 µM), suggesting higher dose of NPs induce genotoxicity [47]. Asharani et al. presented the study of cytotoxicity and genotoxicity of starch-coated silver nanoparticles using normal human lung fibroblast (IMR-90) and human glioblastoma (U251) cells [29]. They noticed an increase in DNA damage with increase in concentration of Ag NP in cancer cells, whereas there was increase in DNA damage only beyond a concentration of 100 µg/mL in the fibroblast cells. A dose-dependent sperm abnormality, clastogenicity and genotoxicity potentiated by Cadmium (Cd) coadministration to Ag NPs were observed [48]. Pesnya reported the absence of induction of chromosomal and mitotic abnormalities by chitosan-capped Ag NPs at low concentration using Allium test [49]. The frequency of chromosomal aberration inversely proportional to the mitotic index (MI) with increased concentration of Ag NPs in dose-dependent manner was demonstrated by Daphedar et al. [50]. They concluded that the Ag NPs synthesized by Getonia floribunda leaf extract cause DNA damage, which leads to cell death at higher concentration of silver NP solution.
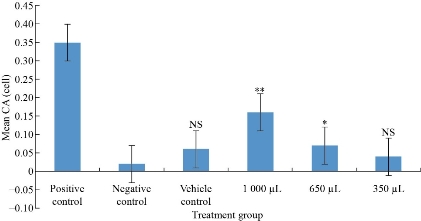
Fig. 7 The mean frequency of chromosomal aberrations in human lymphocytes following in vitro exposure to Ag NPs and controls in short term cultures (**p < 0.001, *p < 0.05, NS statistically not significant).
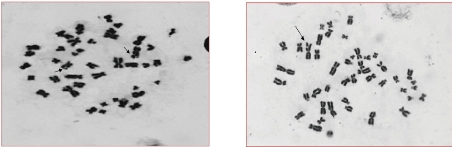
Fig. 8 Representative photomicrographs of Giemsa stained metaphase cells depicting chromosomal aberrations: The short arrows indicates single chromatid gap and the long arrow indicates a single chromatid break (Magnification 100×).
Conclusions
The present study is about characterization and in-vitro biological safety assessment in terms of genotoxicity of green synthesized Ag NPs. The characterization data of Malvern zeta sizer revealed the size of synthesized Ag NPs between 1 to 100 nm and -9.1 mV zeta potential value of Ag NPs suggested their stable form. The morphology and dispersion as well as the presence of elemental silver were studied by TEM and EDS analyses, respectively. To confirm the effect of direct interaction of Ag NPs with the DNA, DNA binding and fluorescence assay was performed. The UV-visible spectroscopy and fluorescence spectroscopy showed no binding of Ag NPs with the DNA, suggesting no direct DNA interaction. Genotoxicity of green synthesized Ag NPs assessed by in-vitro chromosomal aberration assay showed dose dependent increase in frequency of CA as compared to controls (p < 0.05). The primary in-vitro data can be valuable in planning further in-vivo studies for toxicities and possible carcinogenic potential of NPs. These experiments are necessary in view of increasing the use and concern for environmental and cellular toxicity and safety.
Acknowledgements
Authors are grateful to Nirma University for the financial support.
Conflict of Interests
The manuscript has been approved by all the co-authors and declare that no competing interest exists.
References
Copyright© Marcellin Mecwan, Manita Das, Sonal Thakore, and Sonal Rajiv Bakshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.